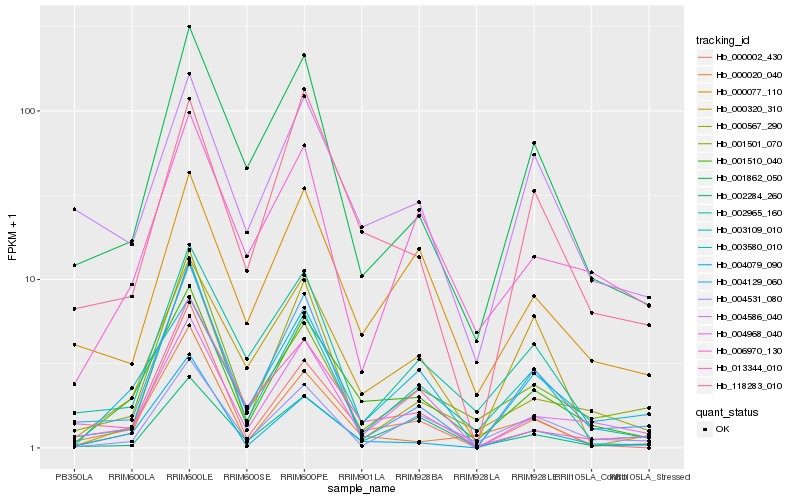
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001510_040 |
0.0 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 2 |
Hb_003580_010 |
0.158048591 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 3 |
Hb_004968_040 |
0.1597463136 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 4 |
Hb_004079_090 |
0.1641545654 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 5 |
Hb_001862_050 |
0.1675578599 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 6 |
Hb_118283_010 |
0.1686340281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003109_010 |
0.1712126526 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 8 |
Hb_000077_110 |
0.1757485789 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 9 |
Hb_002284_260 |
0.1777232851 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 10 |
Hb_000002_430 |
0.1789148806 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 11 |
Hb_004129_060 |
0.1794604271 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_004586_040 |
0.1813590511 |
transcription factor |
TF Family: MIKC |
K-box region and MADS-box transcription factor family protein [Theobroma cacao] |
| 13 |
Hb_000020_040 |
0.182348614 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_002965_160 |
0.1829263636 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 15 |
Hb_004531_080 |
0.1829950978 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 16 |
Hb_006970_130 |
0.1850442717 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 17 |
Hb_000567_290 |
0.1851887751 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 18 |
Hb_013344_010 |
0.1866118645 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 19 |
Hb_001501_070 |
0.1881032168 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 20 |
Hb_000320_310 |
0.1885497174 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |