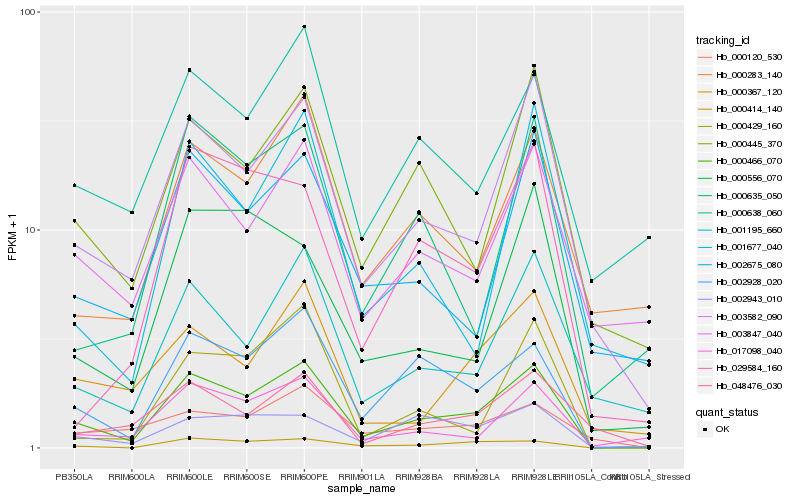
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000466_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 2 |
Hb_002928_020 |
0.130008177 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 3 |
Hb_003582_090 |
0.1365145225 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 4 |
Hb_000414_140 |
0.1585197868 |
- |
- |
Chalcone synthase, putative [Ricinus communis] |
| 5 |
Hb_000445_370 |
0.1721239528 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 6 |
Hb_017098_040 |
0.1721267114 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 7 |
Hb_001195_660 |
0.1793628316 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 8 |
Hb_002943_010 |
0.1851709175 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 9 |
Hb_003847_040 |
0.1855701753 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 10 |
Hb_002675_080 |
0.1881422338 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 11 |
Hb_000367_120 |
0.188788551 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 12 |
Hb_000638_060 |
0.1890847577 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 13 |
Hb_000635_050 |
0.1904732238 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 14 |
Hb_000429_160 |
0.1907846943 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 15 |
Hb_000283_140 |
0.1919083026 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000120_530 |
0.196980525 |
transcription factor |
TF Family: C3H |
RING finger protein 113A, putative [Ricinus communis] |
| 17 |
Hb_000556_070 |
0.1971344946 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 18 |
Hb_029584_160 |
0.1992814934 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 19 |
Hb_001677_040 |
0.2010497679 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 20 |
Hb_048476_030 |
0.2012382006 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |