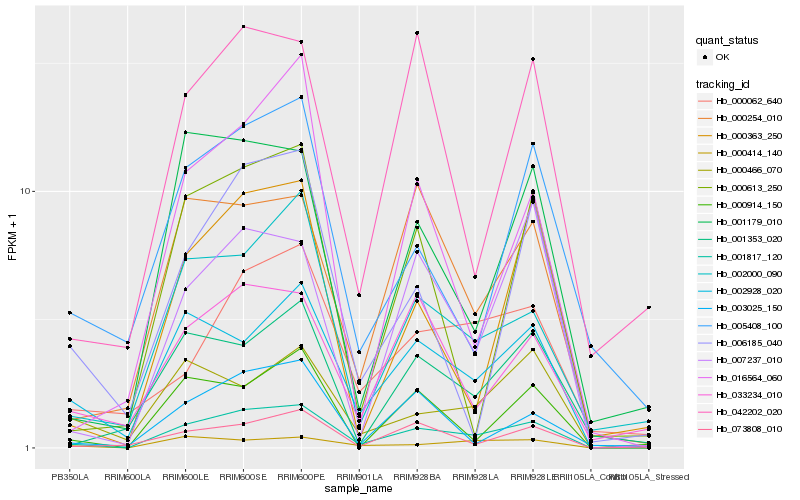
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_120 |
0.0 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_002928_020 |
0.1546127739 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 3 |
Hb_002000_090 |
0.1664490052 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 4 |
Hb_000254_010 |
0.1823299548 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 5 |
Hb_073808_010 |
0.1857595983 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001179_010 |
0.1870216422 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 7 |
Hb_007237_010 |
0.1878688625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_033234_010 |
0.1937744761 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 9 |
Hb_000062_640 |
0.1947493167 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000414_140 |
0.1958960691 |
- |
- |
Chalcone synthase, putative [Ricinus communis] |
| 11 |
Hb_001353_020 |
0.1964741393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000363_250 |
0.197672625 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 13 |
Hb_016564_060 |
0.1983448504 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 14 |
Hb_000914_150 |
0.1996398967 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 15 |
Hb_003025_150 |
0.2006490285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005408_100 |
0.2016487017 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 17 |
Hb_000466_070 |
0.2027720591 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 18 |
Hb_000613_250 |
0.2061548401 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 19 |
Hb_006185_040 |
0.2068721152 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 20 |
Hb_042202_020 |
0.2072786538 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |