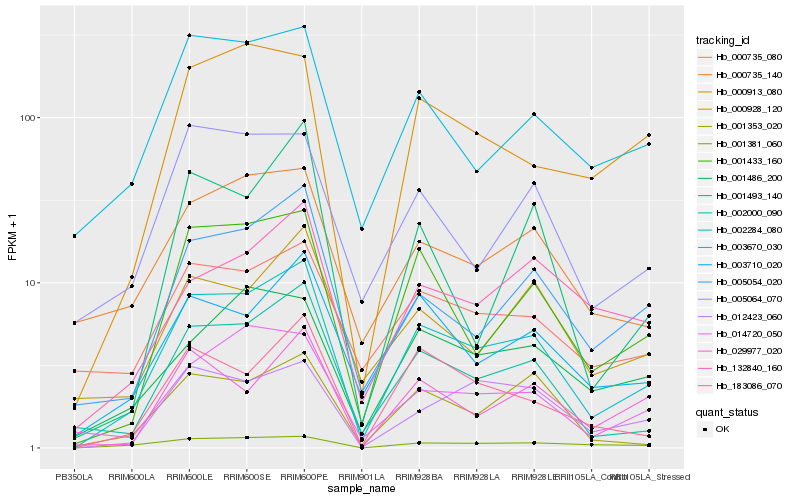
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_080 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_002000_090 |
0.1226582072 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 3 |
Hb_001433_160 |
0.1287054159 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 4 |
Hb_005054_020 |
0.1437538504 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 5 |
Hb_003710_020 |
0.1504884517 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 6 |
Hb_183086_070 |
0.1535101247 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 7 |
Hb_001353_020 |
0.1559367848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001381_060 |
0.159688602 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 9 |
Hb_003670_030 |
0.1597899472 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 10 |
Hb_014720_050 |
0.15979618 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 11 |
Hb_012423_060 |
0.1614881991 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 12 |
Hb_000913_080 |
0.1646661444 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 13 |
Hb_000735_140 |
0.1663951937 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 14 |
Hb_001486_200 |
0.1696393381 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 15 |
Hb_005064_070 |
0.1705473296 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 16 |
Hb_000928_120 |
0.1729525497 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 17 |
Hb_000735_080 |
0.175164827 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 18 |
Hb_029977_020 |
0.1765544816 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001493_140 |
0.1785942775 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 20 |
Hb_132840_160 |
0.1790183688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |