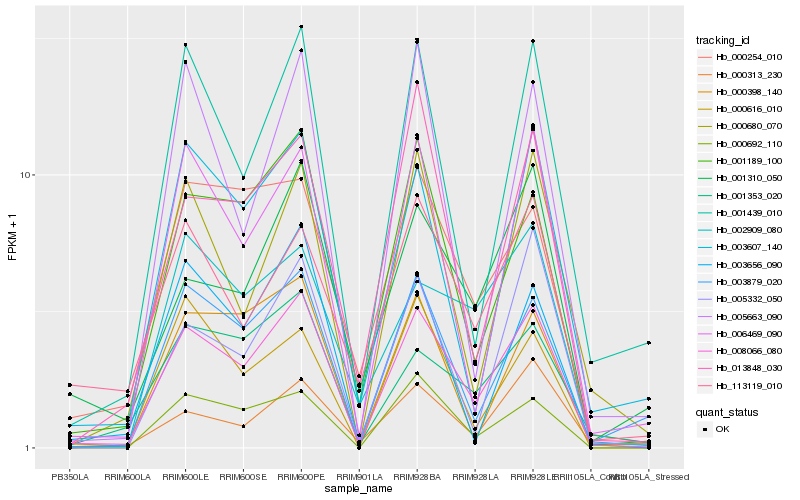
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008066_080 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 2 |
Hb_001189_100 |
0.1191908163 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 3 |
Hb_001439_010 |
0.1301370119 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 4 |
Hb_000692_110 |
0.1329782083 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 5 |
Hb_003879_020 |
0.1338254257 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 6 |
Hb_006469_090 |
0.1349767157 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 7 |
Hb_003607_140 |
0.1452534543 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 8 |
Hb_000680_070 |
0.1487059411 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001353_020 |
0.1515471864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000398_140 |
0.1527698501 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 11 |
Hb_013848_030 |
0.1550000356 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 12 |
Hb_003656_090 |
0.15700666 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 13 |
Hb_005663_090 |
0.1576873668 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 14 |
Hb_000254_010 |
0.1591516396 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 15 |
Hb_001310_050 |
0.1621933391 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 16 |
Hb_000616_010 |
0.1627210746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000313_230 |
0.1630223161 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 18 |
Hb_005332_050 |
0.1639693033 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 19 |
Hb_002909_080 |
0.1656856839 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 20 |
Hb_113119_010 |
0.1666529032 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |