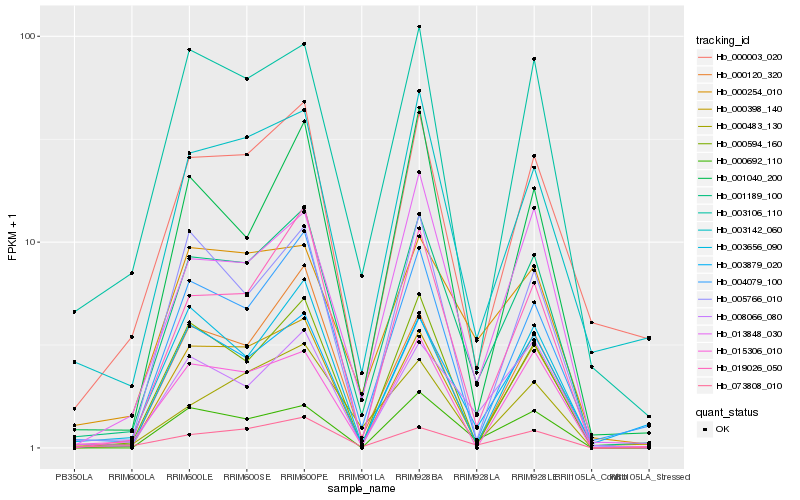
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001189_100 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 2 |
Hb_000398_140 |
0.1007410109 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 3 |
Hb_004079_100 |
0.1126182342 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 4 |
Hb_073808_010 |
0.1152865102 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_019026_050 |
0.1154348121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008066_080 |
0.1191908163 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 7 |
Hb_000594_160 |
0.1217234472 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 8 |
Hb_013848_030 |
0.1224719436 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 9 |
Hb_003142_060 |
0.1227922967 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_003106_110 |
0.1252837698 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000692_110 |
0.1259643885 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 12 |
Hb_003879_020 |
0.1273895687 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 13 |
Hb_000254_010 |
0.130111422 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 14 |
Hb_000003_020 |
0.1310265152 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 15 |
Hb_000120_320 |
0.1349785597 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_001040_200 |
0.137052594 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 17 |
Hb_015306_010 |
0.1378980836 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 18 |
Hb_005766_010 |
0.138242302 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_000483_130 |
0.1404749317 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003656_090 |
0.1416073802 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |