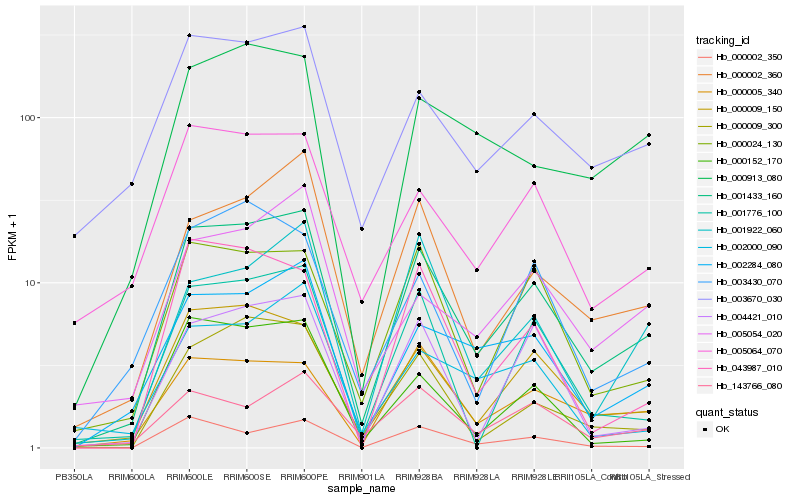
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_160 |
0.0 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 2 |
Hb_000009_150 |
0.108168803 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 3 |
Hb_002284_080 |
0.1287054159 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000002_360 |
0.1314757727 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 5 |
Hb_000009_300 |
0.1408844763 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |
| 6 |
Hb_000024_130 |
0.1426649193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003670_030 |
0.1446150076 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 8 |
Hb_005064_070 |
0.1476710508 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 9 |
Hb_000005_340 |
0.1484479588 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001776_100 |
0.1491154433 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000002_350 |
0.1573638409 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 12 |
Hb_000152_170 |
0.1582591236 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 13 |
Hb_005054_020 |
0.1586401345 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 14 |
Hb_000913_080 |
0.1589189196 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 15 |
Hb_004421_010 |
0.1592128688 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_143766_080 |
0.1603431645 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003430_070 |
0.1622730436 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 18 |
Hb_043987_010 |
0.1625965347 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 19 |
Hb_001922_060 |
0.1657301843 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_002000_090 |
0.1675408503 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |