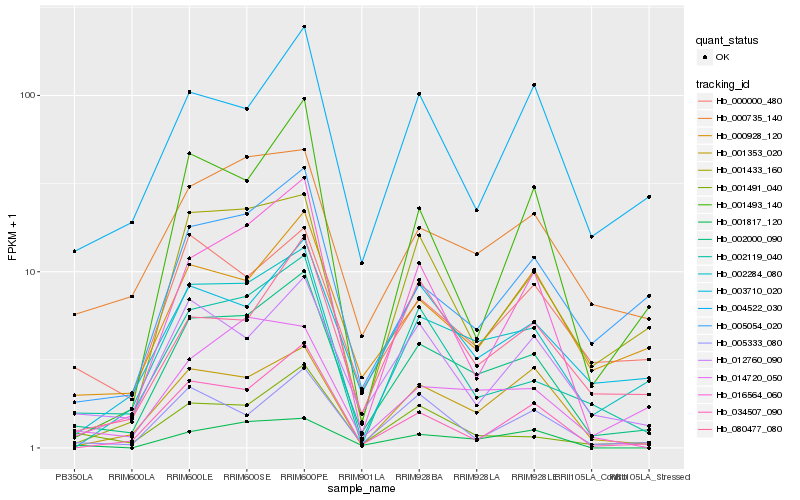
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002000_090 |
0.0 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 2 |
Hb_002284_080 |
0.1226582072 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_001353_020 |
0.1485769898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012760_090 |
0.1487854648 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 5 |
Hb_002119_040 |
0.1511498766 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_034507_090 |
0.1548572454 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000735_140 |
0.1561681797 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 8 |
Hb_016564_060 |
0.1572024398 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 9 |
Hb_005054_020 |
0.1589684061 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 10 |
Hb_001491_040 |
0.1594728731 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 11 |
Hb_003710_020 |
0.160833894 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000928_120 |
0.161969058 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 13 |
Hb_005333_080 |
0.1633294169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001817_120 |
0.1664490052 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_001493_140 |
0.1675210779 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 16 |
Hb_001433_160 |
0.1675408503 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 17 |
Hb_014720_050 |
0.1690946062 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 18 |
Hb_004522_030 |
0.1692856522 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 19 |
Hb_080477_080 |
0.1696636889 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_000000_480 |
0.1700670993 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |