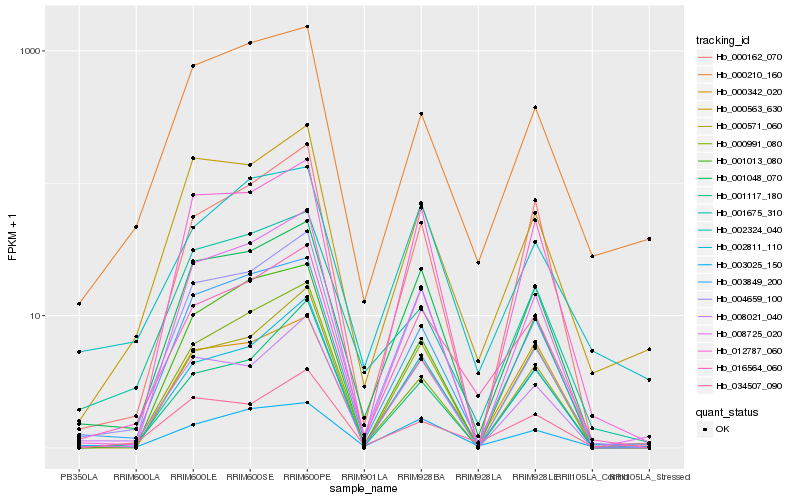
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016564_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 2 |
Hb_001048_070 |
0.1048555796 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 3 |
Hb_003849_200 |
0.1183616282 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_002811_110 |
0.1205036304 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 5 |
Hb_000162_070 |
0.1224321473 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 6 |
Hb_034507_090 |
0.1281516118 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000563_630 |
0.1302908334 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 8 |
Hb_003025_150 |
0.1304134631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008725_020 |
0.1304903567 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 10 |
Hb_000210_160 |
0.1371317274 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 11 |
Hb_000571_060 |
0.1377068446 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 12 |
Hb_012787_060 |
0.1379488546 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 13 |
Hb_000342_020 |
0.1384996826 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 14 |
Hb_008021_040 |
0.1429352117 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 15 |
Hb_001013_080 |
0.1437063246 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 16 |
Hb_001117_180 |
0.1439764696 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000991_080 |
0.1442055913 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_002324_040 |
0.1451715925 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 19 |
Hb_004659_100 |
0.1458454893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001675_310 |
0.1472690726 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |