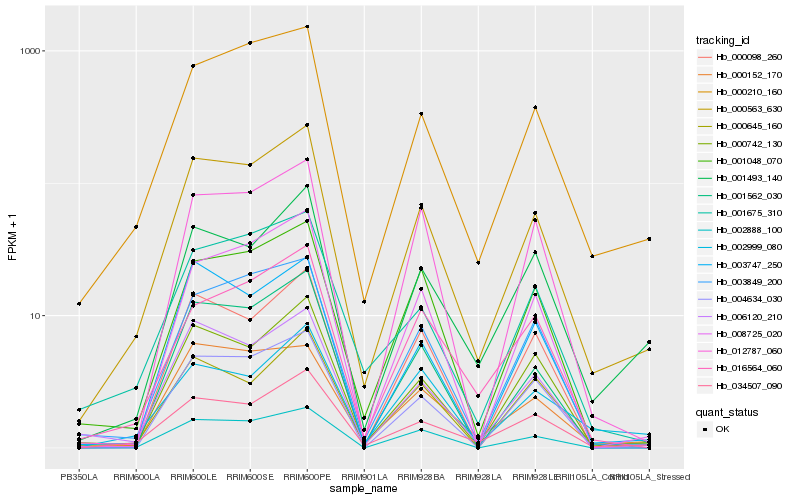
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_630 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 2 |
Hb_034507_090 |
0.0816823081 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000210_160 |
0.088048554 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 4 |
Hb_000742_130 |
0.1079906456 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 5 |
Hb_006120_210 |
0.1096718491 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 6 |
Hb_002888_100 |
0.112241836 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 7 |
Hb_003849_200 |
0.112249994 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_001493_140 |
0.1151370704 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 9 |
Hb_001675_310 |
0.118719538 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001562_030 |
0.1192054643 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008725_020 |
0.1206856235 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 12 |
Hb_000152_170 |
0.1210585129 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 13 |
Hb_001048_070 |
0.1219566493 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 14 |
Hb_000098_260 |
0.124213582 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_000645_160 |
0.1253505122 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 16 |
Hb_003747_250 |
0.130200637 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 17 |
Hb_016564_060 |
0.1302908334 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 18 |
Hb_002999_080 |
0.1315687406 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 19 |
Hb_004634_030 |
0.1331951676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012787_060 |
0.1353316043 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |