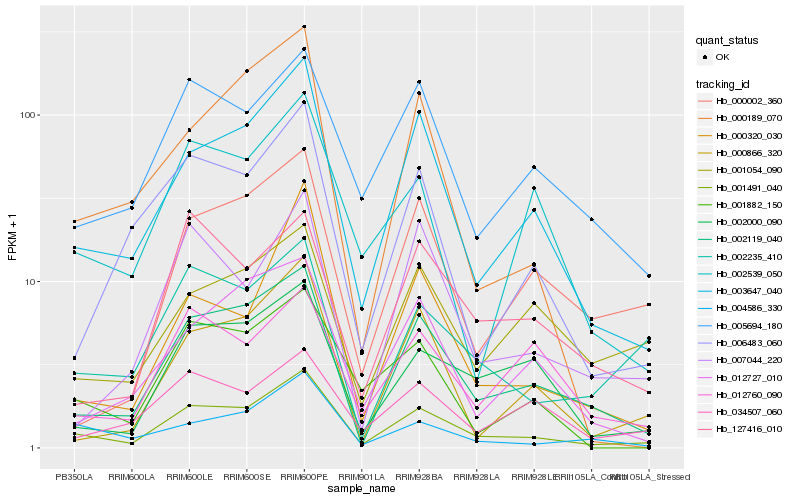
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001491_040 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 2 |
Hb_003647_040 |
0.117188535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002119_040 |
0.1207392512 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_002000_090 |
0.1594728731 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 5 |
Hb_005694_180 |
0.1655752969 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 6 |
Hb_002235_410 |
0.1660119727 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 7 |
Hb_004586_330 |
0.1696022478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000866_320 |
0.1774478875 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_006483_060 |
0.1804740914 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 10 |
Hb_001882_150 |
0.1823845668 |
transcription factor |
TF Family: mTERF |
- |
| 11 |
Hb_002539_050 |
0.1856924779 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 12 |
Hb_001054_090 |
0.1870264174 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 13 |
Hb_000002_360 |
0.1879313675 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 14 |
Hb_012760_090 |
0.1881906935 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 15 |
Hb_000320_030 |
0.1881976436 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 16 |
Hb_007044_220 |
0.1882173876 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_012727_010 |
0.1885188709 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 18 |
Hb_127416_010 |
0.1892726812 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_034507_060 |
0.1919207359 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 20 |
Hb_000189_070 |
0.1928454421 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |