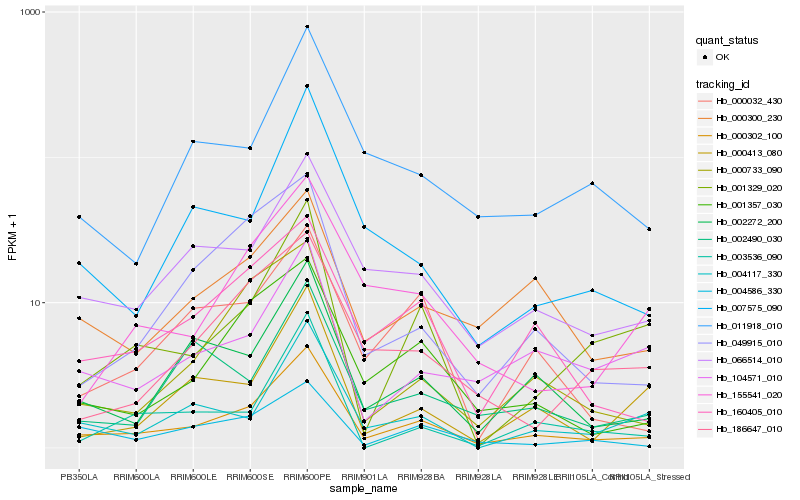
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 2 |
Hb_066514_010 |
0.1483408163 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 3 |
Hb_002272_200 |
0.1575991935 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001357_030 |
0.1606248056 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 5 |
Hb_160405_010 |
0.1640840259 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_007575_090 |
0.1738849866 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 7 |
Hb_001329_020 |
0.17440987 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 8 |
Hb_004117_330 |
0.1764493989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011918_010 |
0.1770640119 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 10 |
Hb_000733_090 |
0.1792644615 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 11 |
Hb_049915_010 |
0.1792653703 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 12 |
Hb_000300_230 |
0.1799336885 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 13 |
Hb_104571_010 |
0.1845885954 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 14 |
Hb_004586_330 |
0.184841838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000032_430 |
0.1852034042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_155541_020 |
0.1863318406 |
- |
- |
- |
| 17 |
Hb_000413_080 |
0.1887255845 |
- |
- |
- |
| 18 |
Hb_186647_010 |
0.192241093 |
- |
- |
- |
| 19 |
Hb_003536_090 |
0.1965855558 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 20 |
Hb_002490_030 |
0.1968593023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |