| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
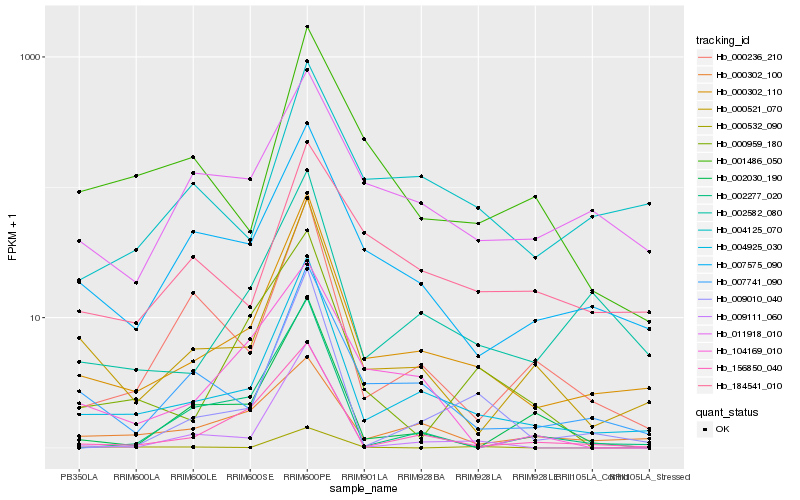
Hb_000302_110 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004925_030 |
0.0693189878 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 3 |
Hb_000521_070 |
0.1509945885 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 4 |
Hb_002582_080 |
0.1550851674 |
- |
- |
- |
| 5 |
Hb_007575_090 |
0.1628890287 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 6 |
Hb_007741_090 |
0.1649717859 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 7 |
Hb_156850_040 |
0.1906069378 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 8 |
Hb_011918_010 |
0.200433625 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 9 |
Hb_000302_100 |
0.2012810626 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 10 |
Hb_009010_040 |
0.2047725701 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 11 |
Hb_004125_070 |
0.2068380975 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 12 |
Hb_000236_210 |
0.2111505731 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 13 |
Hb_001486_050 |
0.2142641342 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 14 |
Hb_000532_090 |
0.218587136 |
- |
- |
ATPase [delta proteobacterium MLMS-1] |
| 15 |
Hb_184541_010 |
0.2201100536 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 16 |
Hb_009111_060 |
0.2202817865 |
- |
- |
PREDICTED: sodium transporter HKT1-like [Jatropha curcas] |
| 17 |
Hb_000959_180 |
0.2208099637 |
- |
- |
hypothetical protein JCGZ_09448 [Jatropha curcas] |
| 18 |
Hb_104169_010 |
0.2260068474 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 19 |
Hb_002030_190 |
0.2267156407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002277_020 |
0.2268882565 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |