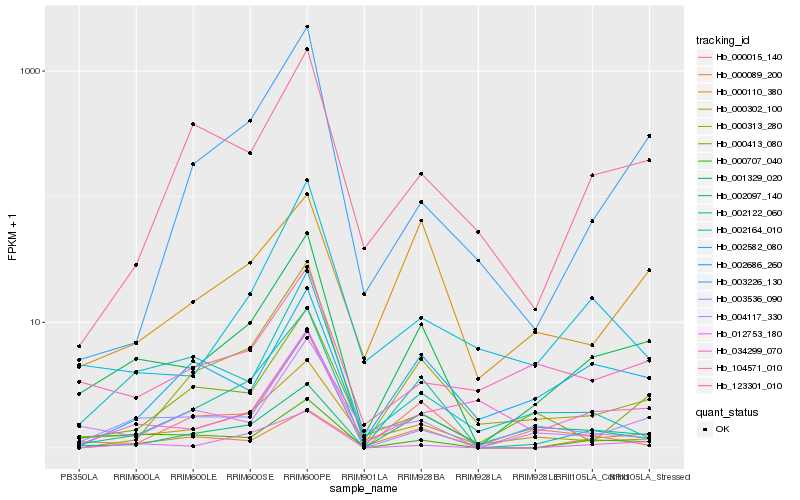
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_020 |
0.0 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 2 |
Hb_000302_100 |
0.17440987 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 3 |
Hb_002122_060 |
0.1942863441 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_004117_330 |
0.2032149966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003536_090 |
0.210034837 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_012753_180 |
0.2110540076 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 7 |
Hb_000707_040 |
0.2130475967 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 8 |
Hb_000313_280 |
0.2161407381 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 9 |
Hb_002582_080 |
0.2211039029 |
- |
- |
- |
| 10 |
Hb_104571_010 |
0.2226689024 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 11 |
Hb_000413_080 |
0.2232873266 |
- |
- |
- |
| 12 |
Hb_000089_200 |
0.2301237093 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 13 |
Hb_000110_380 |
0.2313703126 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 14 |
Hb_002686_260 |
0.2319533749 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 15 |
Hb_123301_010 |
0.236487702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003226_130 |
0.2433360075 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 17 |
Hb_034299_070 |
0.244577717 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 18 |
Hb_002097_140 |
0.2450561757 |
- |
- |
CHY zinc finger family protein, expressed [Oryza sativa Japonica Group] |
| 19 |
Hb_002164_010 |
0.2470196746 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 20 |
Hb_000015_140 |
0.2501827655 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |