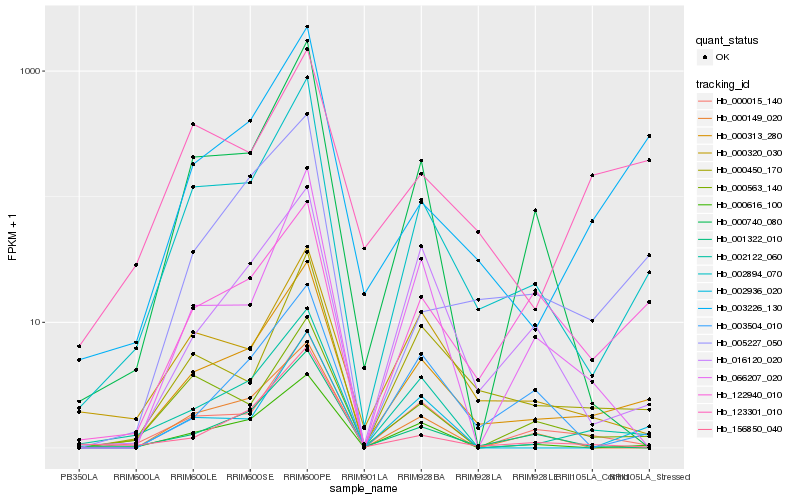
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_280 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 2 |
Hb_002894_070 |
0.1010257473 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 3 |
Hb_002122_060 |
0.1421166145 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000015_140 |
0.1522706897 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 5 |
Hb_000616_100 |
0.1525673193 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000450_170 |
0.1601924882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003226_130 |
0.1725238749 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 8 |
Hb_001322_010 |
0.1795696588 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 9 |
Hb_005227_050 |
0.1857466928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000563_140 |
0.1861103618 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 11 |
Hb_000740_080 |
0.1874310471 |
- |
- |
- |
| 12 |
Hb_000320_030 |
0.1888721705 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 13 |
Hb_002936_020 |
0.1898216365 |
- |
- |
hypothetical protein VITISV_001748 [Vitis vinifera] |
| 14 |
Hb_156850_040 |
0.191374305 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 15 |
Hb_016120_020 |
0.1935717069 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |
| 16 |
Hb_122940_010 |
0.1951471177 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_123301_010 |
0.1978967838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_066207_020 |
0.1988612562 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 19 |
Hb_000149_020 |
0.1988788185 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 20 |
Hb_003504_010 |
0.2001565568 |
- |
- |
hypothetical protein POPTR_0013s04290g [Populus trichocarpa] |