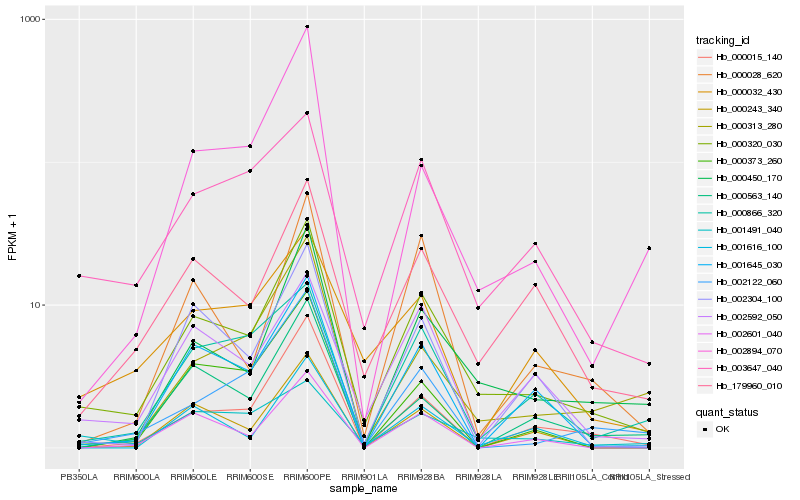
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_030 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 2 |
Hb_000450_170 |
0.1513358458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_179960_010 |
0.159280755 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 4 |
Hb_003647_040 |
0.1756019803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000015_140 |
0.176736912 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 6 |
Hb_002592_050 |
0.1777664053 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 7 |
Hb_001645_030 |
0.1811852119 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1b [Jatropha curcas] |
| 8 |
Hb_000028_620 |
0.1859179164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002894_070 |
0.1859743134 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 10 |
Hb_002304_100 |
0.1863083674 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 11 |
Hb_002122_060 |
0.1871479446 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_001491_040 |
0.1881976436 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_000313_280 |
0.1888721705 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 14 |
Hb_000243_340 |
0.1897121632 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 15 |
Hb_001616_100 |
0.1917096949 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 16 |
Hb_000866_320 |
0.1924614268 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000563_140 |
0.1925844631 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_002601_040 |
0.192793997 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |
| 19 |
Hb_000373_260 |
0.1930316232 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 20 |
Hb_000032_430 |
0.1932130007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |