| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
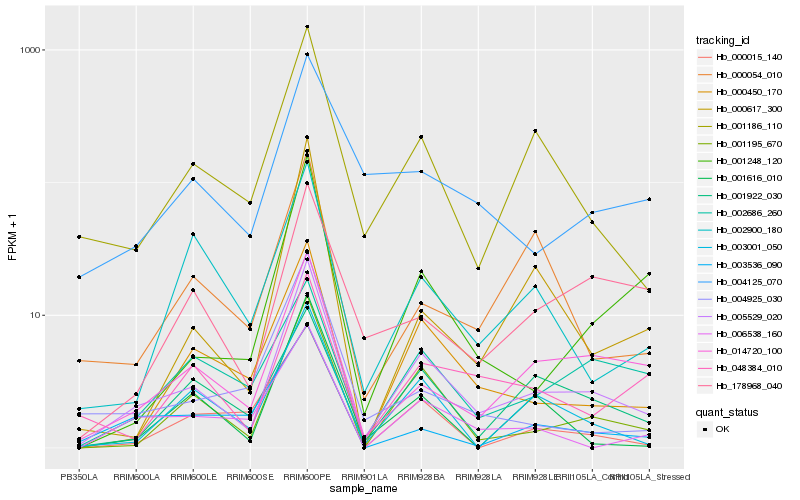
Hb_005529_020 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 2 |
Hb_001195_670 |
0.1577648504 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 3 |
Hb_000450_170 |
0.1779540559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001186_110 |
0.1792150243 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 5 |
Hb_014720_100 |
0.1809520928 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 6 |
Hb_003001_050 |
0.1902171461 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 7 |
Hb_006538_160 |
0.2066508764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001248_120 |
0.2114957332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001922_030 |
0.2124412844 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 10 |
Hb_001616_010 |
0.2154285702 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 11 |
Hb_048384_010 |
0.2157394985 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g42960 [Cucumis sativus] |
| 12 |
Hb_000617_300 |
0.2165281103 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 13 |
Hb_004125_070 |
0.2181597909 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 14 |
Hb_000015_140 |
0.2212384461 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 15 |
Hb_002686_260 |
0.2222155952 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 16 |
Hb_002900_180 |
0.2230726826 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 17 |
Hb_003536_090 |
0.2272787612 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 18 |
Hb_178968_040 |
0.2281771824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004925_030 |
0.2281938064 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 20 |
Hb_000054_010 |
0.2287267461 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |