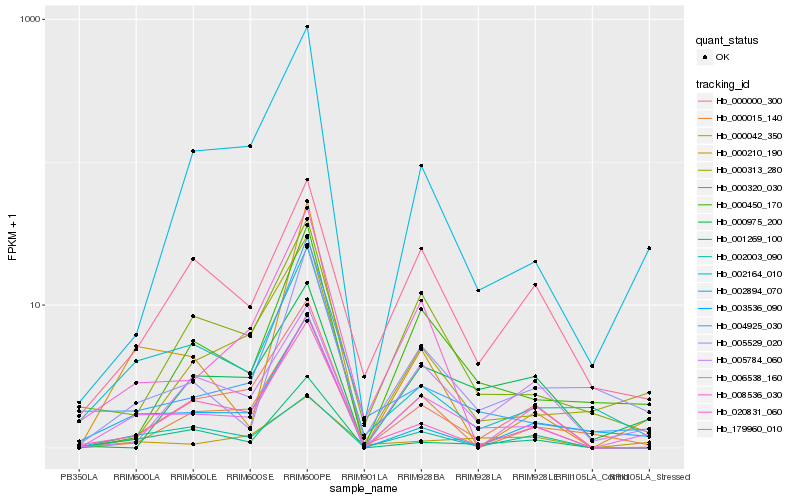
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000450_170 |
0.1911234824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000000_300 |
0.1992461806 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 4 |
Hb_002894_070 |
0.2017578122 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 5 |
Hb_001269_100 |
0.2038902609 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |
| 6 |
Hb_005529_020 |
0.2066508764 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 7 |
Hb_003536_090 |
0.2112467975 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 8 |
Hb_020831_060 |
0.2144838892 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 9 |
Hb_000313_280 |
0.2168089558 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 10 |
Hb_000320_030 |
0.2171479651 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 11 |
Hb_000210_190 |
0.2182851888 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 12 |
Hb_002164_010 |
0.2183879672 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 13 |
Hb_000042_350 |
0.2234143291 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 14 |
Hb_000015_140 |
0.2235679127 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 15 |
Hb_179960_010 |
0.2249578752 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 16 |
Hb_008536_030 |
0.2259484422 |
- |
- |
hypothetical protein POPTR_0005s08290g [Populus trichocarpa] |
| 17 |
Hb_004925_030 |
0.2280926046 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 18 |
Hb_002003_090 |
0.2291969569 |
- |
- |
PREDICTED: TPR repeat-containing protein ZIP4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000975_200 |
0.2291974575 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005784_060 |
0.2328114436 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |