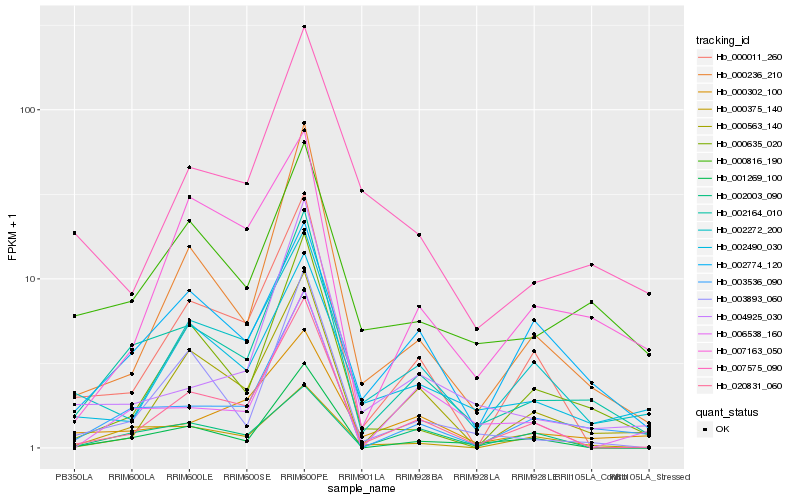
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002164_010 |
0.0 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 2 |
Hb_003536_090 |
0.1220566853 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_000236_210 |
0.1612466503 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 4 |
Hb_000375_140 |
0.1654630104 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 5 |
Hb_001269_100 |
0.1751004986 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |
| 6 |
Hb_020831_060 |
0.1752465245 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 7 |
Hb_000816_190 |
0.1824307178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003893_060 |
0.1853807157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000635_020 |
0.1914916368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000011_260 |
0.1962425663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000302_100 |
0.1970101024 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 12 |
Hb_004925_030 |
0.2044302296 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 13 |
Hb_002774_120 |
0.2079810597 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 14 |
Hb_002490_030 |
0.2093304805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002272_200 |
0.2101351223 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000563_140 |
0.2119835151 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 17 |
Hb_007163_050 |
0.2137932158 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 18 |
Hb_007575_090 |
0.2138162491 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 19 |
Hb_002003_090 |
0.2174665249 |
- |
- |
PREDICTED: TPR repeat-containing protein ZIP4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006538_160 |
0.2183879672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |