| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
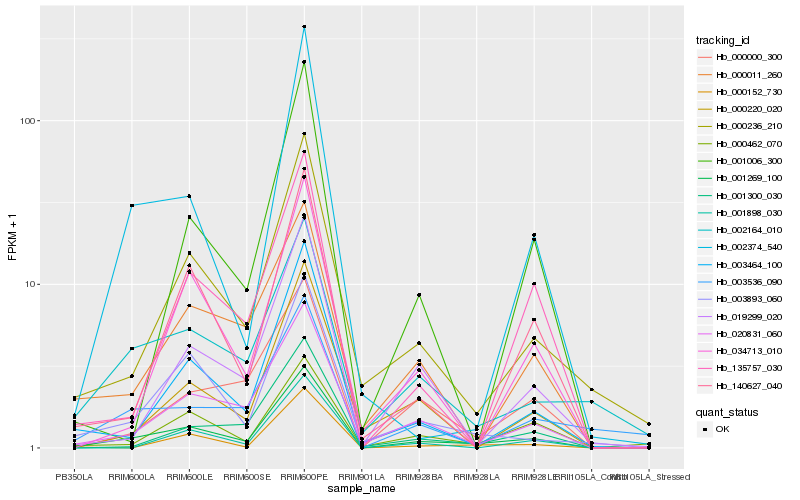
Hb_001269_100 |
0.0 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |
| 2 |
Hb_020831_060 |
0.1429017148 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 3 |
Hb_000236_210 |
0.1579443078 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 4 |
Hb_003893_060 |
0.1602546212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003464_100 |
0.1623879099 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 6 |
Hb_002164_010 |
0.1751004986 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 7 |
Hb_140627_040 |
0.181120112 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 8 |
Hb_000462_070 |
0.1816680984 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 9 |
Hb_019299_020 |
0.1833769679 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 10 |
Hb_000011_260 |
0.1849427669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_034713_010 |
0.1861940626 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 12 |
Hb_000220_020 |
0.1869270434 |
- |
- |
hypothetical protein CICLE_v10017575mg [Citrus clementina] |
| 13 |
Hb_001300_030 |
0.1869717348 |
- |
- |
hypothetical protein JCGZ_13871 [Jatropha curcas] |
| 14 |
Hb_000000_300 |
0.1878467581 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 15 |
Hb_135757_030 |
0.1883837868 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 16 |
Hb_000152_730 |
0.1903599707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003536_090 |
0.1955154282 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 18 |
Hb_002374_540 |
0.196336225 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 19 |
Hb_001006_300 |
0.1981078278 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_001898_030 |
0.198932437 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |