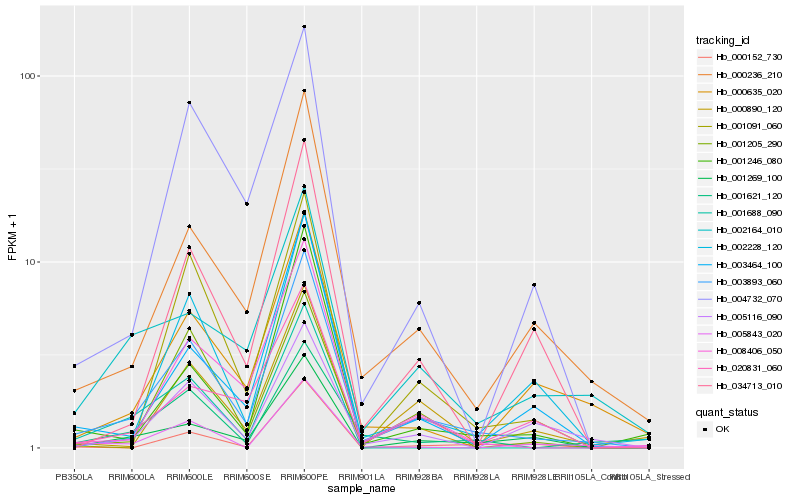
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003893_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000236_210 |
0.1342942212 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 3 |
Hb_008406_050 |
0.1560439251 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_003464_100 |
0.1589602019 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 5 |
Hb_001269_100 |
0.1602546212 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |
| 6 |
Hb_004732_070 |
0.1721440508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001246_080 |
0.1751485015 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 8 |
Hb_002228_120 |
0.177519861 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 9 |
Hb_005116_090 |
0.1780572779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000890_120 |
0.1812540982 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001091_060 |
0.1830523368 |
- |
- |
hypothetical protein JCGZ_27062 [Jatropha curcas] |
| 12 |
Hb_020831_060 |
0.1850509499 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 13 |
Hb_002164_010 |
0.1853807157 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 14 |
Hb_005843_020 |
0.1858143599 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 15 |
Hb_000635_020 |
0.1891862028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_034713_010 |
0.1896215994 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 17 |
Hb_000152_730 |
0.1914843223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001688_090 |
0.1935286967 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 19 |
Hb_001205_290 |
0.1935368855 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 20 |
Hb_001621_120 |
0.1944943821 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |