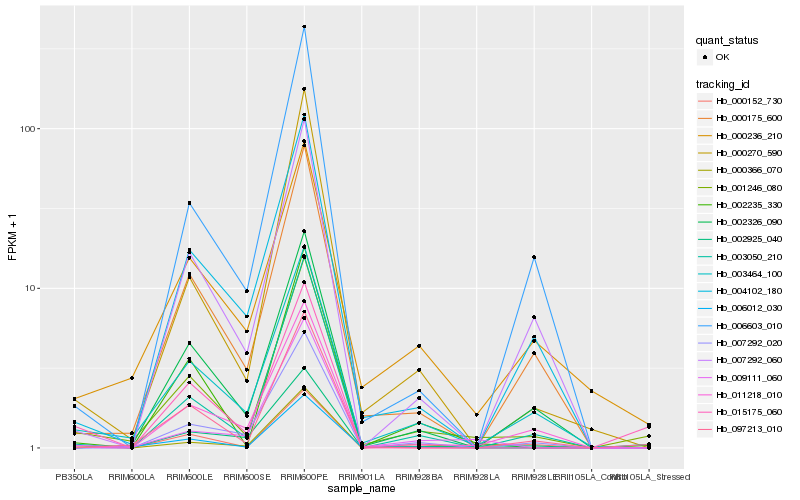
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001246_080 |
0.0 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 2 |
Hb_003464_100 |
0.1269489521 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 3 |
Hb_003050_210 |
0.1306260298 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 4 |
Hb_000152_730 |
0.1355305065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002235_330 |
0.137133021 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_097213_010 |
0.1426750141 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 7 |
Hb_000175_600 |
0.1454675043 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 8 |
Hb_000270_590 |
0.1480302751 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 9 |
Hb_002925_040 |
0.1557564458 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 10 |
Hb_002326_090 |
0.1575630552 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_004102_180 |
0.1596995228 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 12 |
Hb_006012_030 |
0.1603778477 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 13 |
Hb_011218_010 |
0.1611537219 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 14 |
Hb_009111_060 |
0.1640879301 |
- |
- |
PREDICTED: sodium transporter HKT1-like [Jatropha curcas] |
| 15 |
Hb_007292_060 |
0.164500876 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 16 |
Hb_000236_210 |
0.164827152 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 17 |
Hb_006603_010 |
0.1680739078 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 18 |
Hb_015175_060 |
0.1683711771 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 19 |
Hb_000366_070 |
0.1712598044 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.2 [Jatropha curcas] |
| 20 |
Hb_007292_020 |
0.1722433462 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |