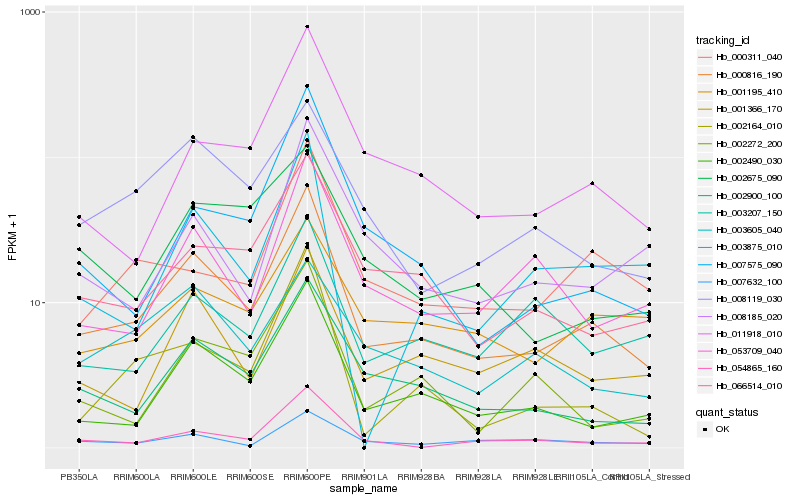
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002490_030 |
0.1337783717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002900_100 |
0.1391510681 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 4 |
Hb_066514_010 |
0.1532061282 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 5 |
Hb_008119_030 |
0.1633174509 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 6 |
Hb_001366_170 |
0.1664444045 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 7 |
Hb_011918_010 |
0.1669434572 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 8 |
Hb_054865_160 |
0.1674070866 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 9 |
Hb_053709_040 |
0.1702030468 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 10 |
Hb_000311_040 |
0.1765002389 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 11 |
Hb_001195_410 |
0.1795008526 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 12 |
Hb_003605_040 |
0.1809808542 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008185_020 |
0.181525331 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 14 |
Hb_002164_010 |
0.1824307178 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 15 |
Hb_002272_200 |
0.1832496672 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002675_090 |
0.1835008464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007575_090 |
0.1838432299 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 18 |
Hb_003875_010 |
0.1853542903 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 19 |
Hb_003207_150 |
0.1875535598 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 20 |
Hb_007632_100 |
0.1875907644 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |