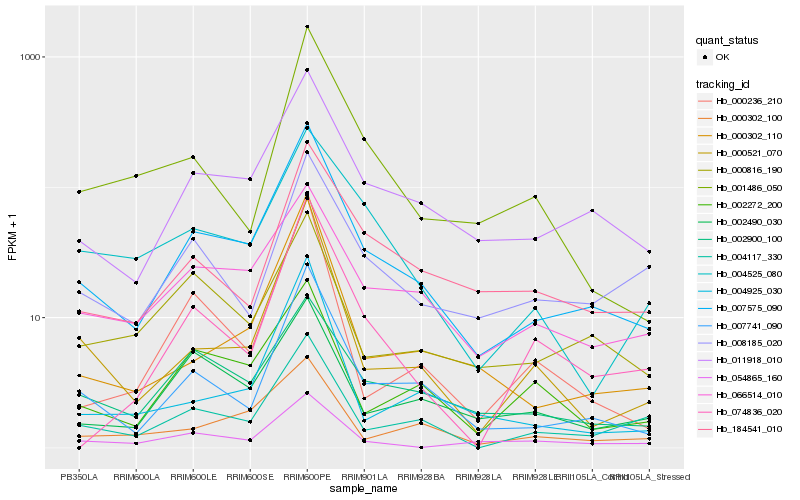
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007575_090 |
0.0 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 2 |
Hb_011918_010 |
0.1077357776 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 3 |
Hb_007741_090 |
0.1169336243 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 4 |
Hb_066514_010 |
0.1507328233 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 5 |
Hb_002900_100 |
0.1577790724 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 6 |
Hb_000302_110 |
0.1628890287 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_004117_330 |
0.1636554177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000521_070 |
0.1648152025 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 9 |
Hb_184541_010 |
0.1661441402 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 10 |
Hb_002490_030 |
0.1689130626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002272_200 |
0.1695412588 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004525_080 |
0.1711322733 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 13 |
Hb_000302_100 |
0.1738849866 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 14 |
Hb_054865_160 |
0.181967401 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 15 |
Hb_008185_020 |
0.1821717485 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 16 |
Hb_000816_190 |
0.1838432299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001486_050 |
0.1844730591 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 18 |
Hb_000236_210 |
0.1871899499 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 19 |
Hb_004925_030 |
0.1872252616 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 20 |
Hb_074836_020 |
0.1941230981 |
- |
- |
unnamed protein product [Vitis vinifera] |