| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
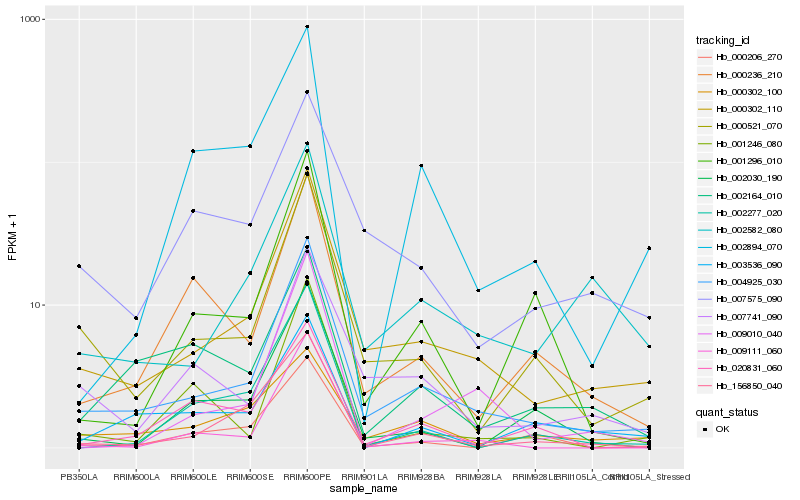
Hb_004925_030 |
0.0 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 2 |
Hb_000302_110 |
0.0693189878 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000521_070 |
0.137223288 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 4 |
Hb_007741_090 |
0.1722893896 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 5 |
Hb_002582_080 |
0.1784856012 |
- |
- |
- |
| 6 |
Hb_156850_040 |
0.1790250665 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 7 |
Hb_000236_210 |
0.1831106467 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 8 |
Hb_007575_090 |
0.1872252616 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 9 |
Hb_009010_040 |
0.1972054549 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 10 |
Hb_009111_060 |
0.1988316981 |
- |
- |
PREDICTED: sodium transporter HKT1-like [Jatropha curcas] |
| 11 |
Hb_001296_010 |
0.2026106515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000302_100 |
0.2026488418 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 13 |
Hb_002164_010 |
0.2044302296 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 14 |
Hb_002030_190 |
0.2058030212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000206_270 |
0.2059587425 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_003536_090 |
0.2073469 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_002277_020 |
0.2086107984 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 18 |
Hb_020831_060 |
0.2087004117 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 19 |
Hb_001246_080 |
0.2095647843 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 20 |
Hb_002894_070 |
0.2110754358 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |