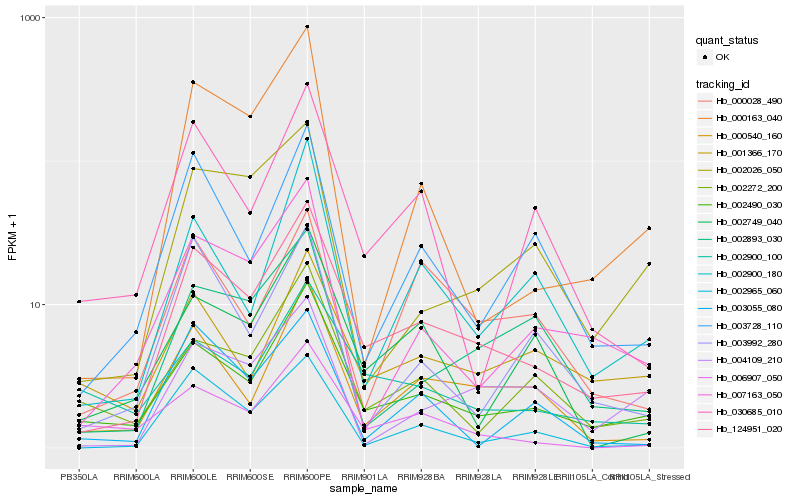
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124951_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002490_030 |
0.1259566314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000540_160 |
0.1600636784 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 4 |
Hb_002965_060 |
0.1741305448 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 5 |
Hb_003728_110 |
0.1822635167 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 6 |
Hb_007163_050 |
0.1839000985 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 7 |
Hb_030685_010 |
0.1847580181 |
- |
- |
- |
| 8 |
Hb_002900_100 |
0.1910704877 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 9 |
Hb_000163_040 |
0.1942066037 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 10 |
Hb_002272_200 |
0.1944918909 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002900_180 |
0.196390578 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 12 |
Hb_002893_030 |
0.1991294797 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 13 |
Hb_002749_040 |
0.1993920043 |
- |
- |
Ribosomal protein L6 family protein [Theobroma cacao] |
| 14 |
Hb_006907_050 |
0.2035339911 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_001366_170 |
0.204829128 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 16 |
Hb_002026_050 |
0.2082892751 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 17 |
Hb_003055_080 |
0.20838964 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000028_490 |
0.2089225566 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 19 |
Hb_004109_210 |
0.2094931134 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_003992_280 |
0.210202168 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |