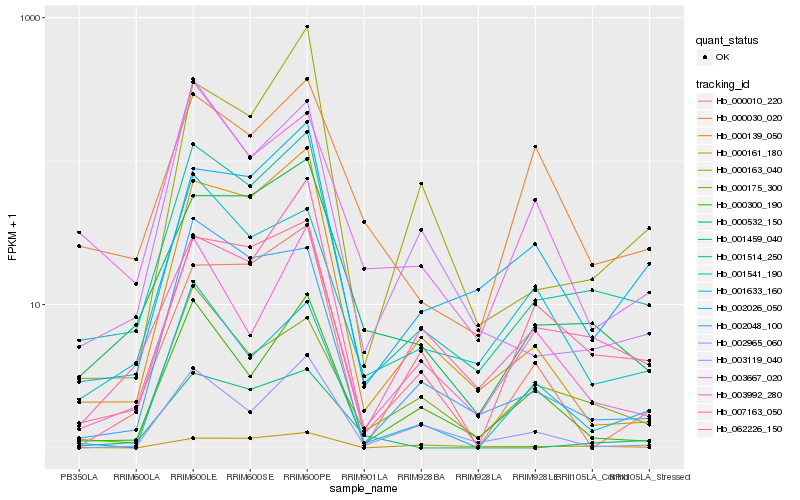
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_250 |
0.0 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 2 |
Hb_000532_150 |
0.1347153499 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 3 |
Hb_007163_050 |
0.1622553959 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 4 |
Hb_000139_050 |
0.1649855247 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 5 |
Hb_062226_150 |
0.167962576 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002026_050 |
0.1720533076 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 7 |
Hb_000300_190 |
0.1821428099 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 8 |
Hb_002048_100 |
0.1843143642 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 9 |
Hb_003119_040 |
0.1850255 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 10 |
Hb_001541_190 |
0.1857106733 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 11 |
Hb_003992_280 |
0.1883811381 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 12 |
Hb_001633_160 |
0.1920252206 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001459_040 |
0.1946907303 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 14 |
Hb_000161_180 |
0.195247334 |
- |
- |
Protein argonaute 2 [Morus notabilis] |
| 15 |
Hb_000163_040 |
0.1952825518 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 16 |
Hb_000175_300 |
0.1955531338 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 17 |
Hb_002965_060 |
0.1981829634 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 18 |
Hb_003667_020 |
0.198477702 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 19 |
Hb_000030_020 |
0.1994418958 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 20 |
Hb_000010_220 |
0.201424566 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |