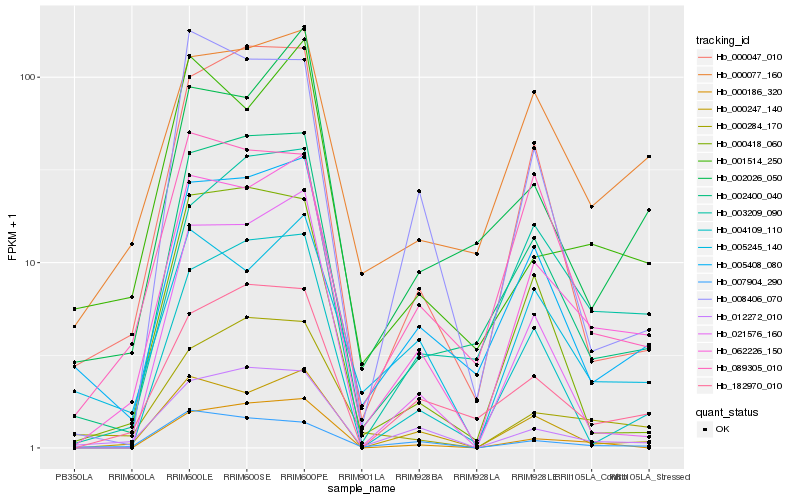
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062226_150 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000247_140 |
0.1069700656 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 3 |
Hb_005408_080 |
0.1427066518 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 4 |
Hb_002400_040 |
0.1442397065 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 5 |
Hb_012272_010 |
0.1553515712 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_182970_010 |
0.159828099 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 7 |
Hb_021576_160 |
0.159915152 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_003209_090 |
0.1607794912 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 9 |
Hb_000077_160 |
0.1625744558 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 10 |
Hb_005245_140 |
0.163574957 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 11 |
Hb_000047_010 |
0.1637190061 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 12 |
Hb_000186_320 |
0.1678748481 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_001514_250 |
0.167962576 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 14 |
Hb_004109_110 |
0.1696226834 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 15 |
Hb_007904_290 |
0.1712480972 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 16 |
Hb_002026_050 |
0.1712599467 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 17 |
Hb_000284_170 |
0.1743443679 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_089305_010 |
0.1780637384 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 19 |
Hb_000418_060 |
0.1786293742 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 20 |
Hb_008406_070 |
0.1798503727 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |