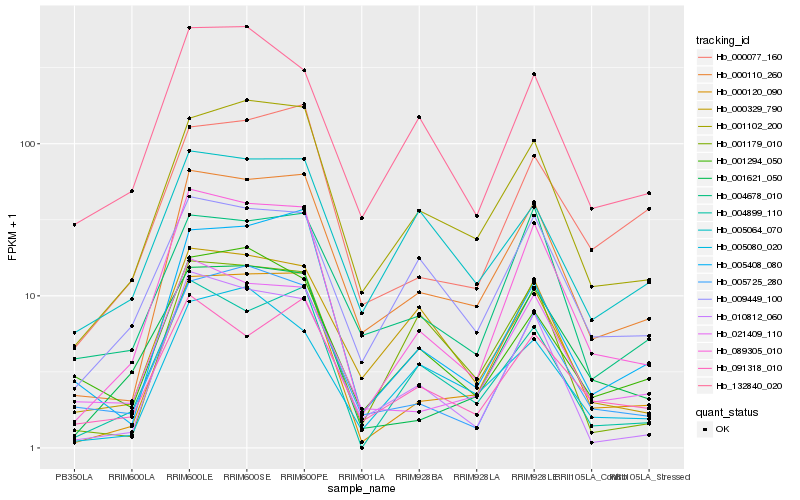
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 2 |
Hb_001102_200 |
0.0902981818 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 3 |
Hb_089305_010 |
0.110965745 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 4 |
Hb_000120_090 |
0.1178777068 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 5 |
Hb_021409_110 |
0.1240907742 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_005408_080 |
0.1295509583 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 7 |
Hb_091318_010 |
0.1302997444 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 8 |
Hb_010812_060 |
0.1338319665 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 9 |
Hb_000329_790 |
0.1350024248 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 10 |
Hb_004678_010 |
0.1360405185 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 11 |
Hb_000077_160 |
0.1367379957 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 12 |
Hb_005725_280 |
0.1386283526 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 13 |
Hb_009449_100 |
0.1436223191 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 14 |
Hb_005064_070 |
0.1455706258 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 15 |
Hb_001179_010 |
0.1461368938 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 16 |
Hb_004899_110 |
0.1477736931 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 17 |
Hb_001294_050 |
0.151193859 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 18 |
Hb_005080_020 |
0.1515391206 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 19 |
Hb_001621_050 |
0.1534996665 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 20 |
Hb_132840_020 |
0.1557809691 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |