| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
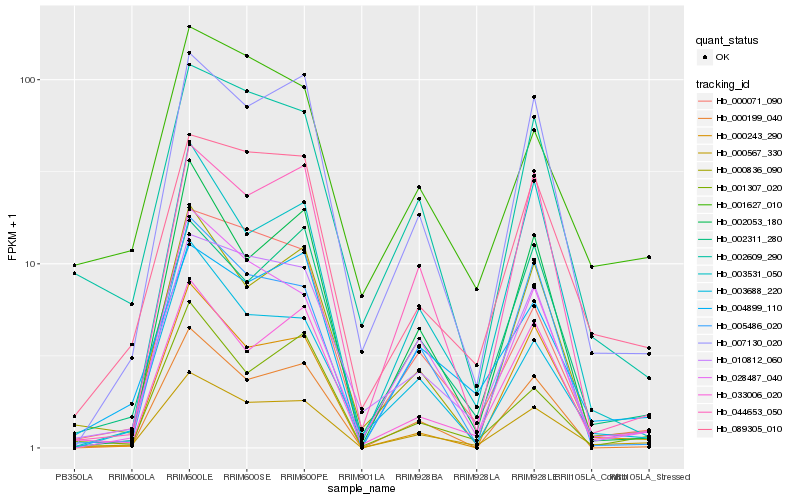
Hb_000567_330 |
0.0 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 2 |
Hb_089305_010 |
0.1088926482 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 3 |
Hb_007130_020 |
0.1169294616 |
- |
- |
- |
| 4 |
Hb_033006_020 |
0.117708323 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_004899_110 |
0.1210127379 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 6 |
Hb_000836_090 |
0.1240926635 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 7 |
Hb_010812_060 |
0.1245373349 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 8 |
Hb_028487_040 |
0.1292319332 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 9 |
Hb_002311_280 |
0.1322025118 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 10 |
Hb_002609_290 |
0.1329656384 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 11 |
Hb_000071_090 |
0.1363426508 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 12 |
Hb_003688_220 |
0.1364701471 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_000199_040 |
0.1368524331 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005486_020 |
0.1409925026 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 15 |
Hb_000243_290 |
0.1411861894 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |
| 16 |
Hb_001627_010 |
0.1416352348 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 17 |
Hb_001307_020 |
0.1426115982 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 18 |
Hb_044653_050 |
0.1432794756 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002053_180 |
0.1443935278 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 20 |
Hb_003531_050 |
0.1444530532 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |