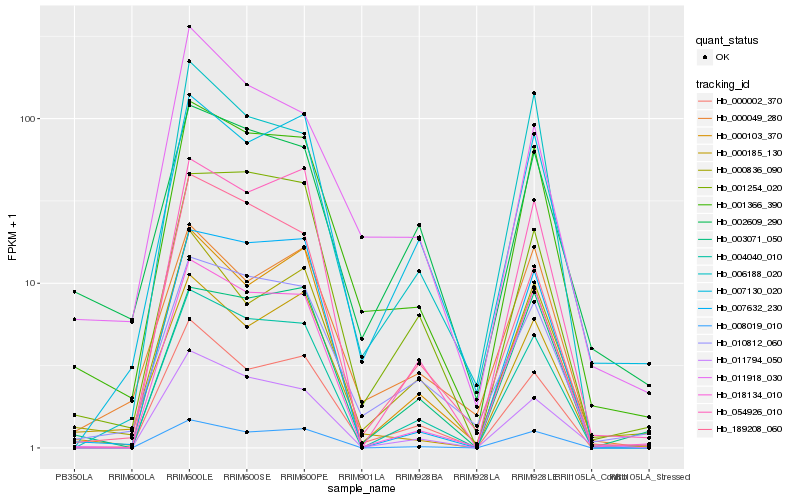
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 2 |
Hb_010812_060 |
0.0943991459 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 3 |
Hb_004040_010 |
0.1148821406 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 4 |
Hb_000002_370 |
0.1151606712 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 5 |
Hb_018134_010 |
0.1154106376 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011794_050 |
0.1164194067 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000185_130 |
0.1204103951 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |
| 8 |
Hb_007130_020 |
0.1227480183 |
- |
- |
- |
| 9 |
Hb_000836_090 |
0.1228012156 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 10 |
Hb_001254_020 |
0.1254714227 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 11 |
Hb_008019_010 |
0.128158075 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 12 |
Hb_000103_370 |
0.1282462278 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 13 |
Hb_054926_010 |
0.1321771173 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 14 |
Hb_000049_280 |
0.1340062503 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 15 |
Hb_011918_030 |
0.1358863513 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 16 |
Hb_006188_020 |
0.1370024108 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 17 |
Hb_002609_290 |
0.1372355072 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 18 |
Hb_007632_230 |
0.1393492268 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 19 |
Hb_003071_050 |
0.1397163901 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 20 |
Hb_189208_060 |
0.1413443964 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |