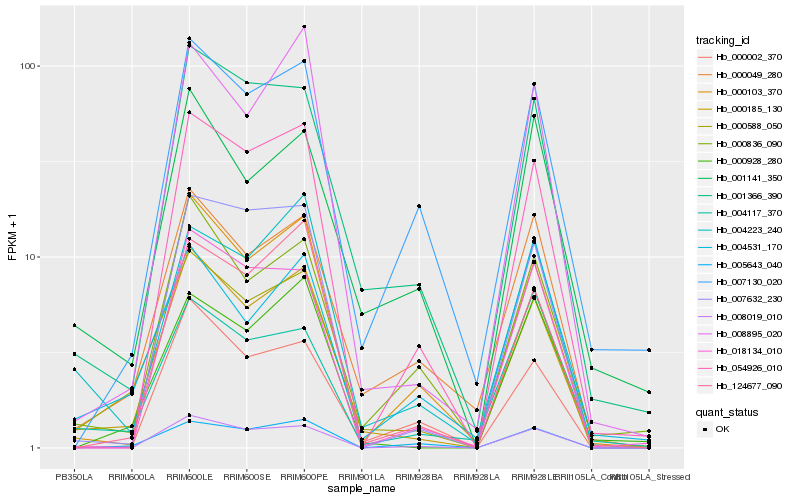
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000588_050 |
0.0 |
transcription factor |
TF Family: Trihelix |
hypothetical protein POPTR_0008s02580g [Populus trichocarpa] |
| 2 |
Hb_000185_130 |
0.0838561247 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |
| 3 |
Hb_000049_280 |
0.1252433765 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 4 |
Hb_001366_390 |
0.1422380271 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 5 |
Hb_005643_040 |
0.1446271115 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000103_370 |
0.1559710739 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 7 |
Hb_000928_280 |
0.1560378027 |
- |
- |
hypothetical protein VITISV_005300 [Vitis vinifera] |
| 8 |
Hb_000836_090 |
0.1566475786 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 9 |
Hb_004531_170 |
0.1611511319 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 10 |
Hb_001141_350 |
0.1632627366 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 11 |
Hb_008019_010 |
0.1632915209 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 12 |
Hb_008895_020 |
0.1647922828 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 13 |
Hb_000002_370 |
0.1661965669 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 14 |
Hb_124677_090 |
0.1666695532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004223_240 |
0.1677123921 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 16 |
Hb_018134_010 |
0.1681889057 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 17 |
Hb_054926_010 |
0.1682087898 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 18 |
Hb_004117_370 |
0.1685348442 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 19 |
Hb_007130_020 |
0.1688814393 |
- |
- |
- |
| 20 |
Hb_007632_230 |
0.1701841898 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |