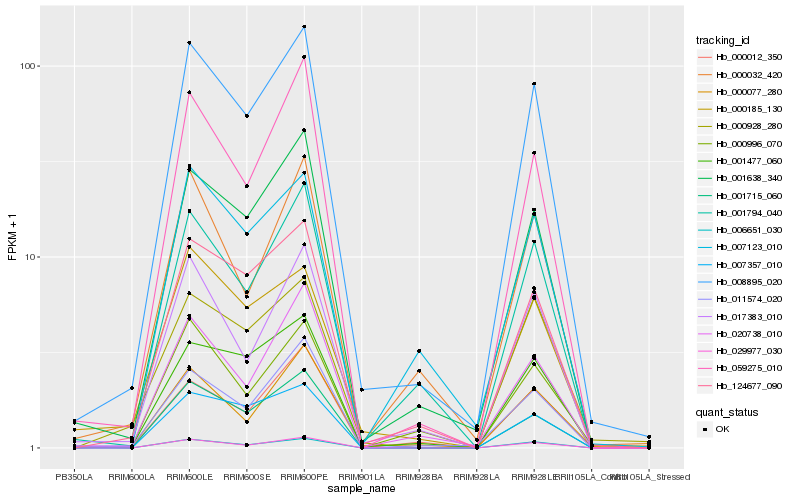
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008895_020 |
0.0 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 2 |
Hb_001638_340 |
0.0794364703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006651_030 |
0.0828017662 |
- |
- |
unnamed protein product [Homo sapiens] |
| 4 |
Hb_029977_030 |
0.0833990935 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_124677_090 |
0.0837331204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001477_060 |
0.0973621813 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_059275_010 |
0.0981142921 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 8 |
Hb_001794_040 |
0.0981802594 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 9 |
Hb_017383_010 |
0.0991401678 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 10 |
Hb_001715_060 |
0.1012227252 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 11 |
Hb_011574_020 |
0.1015563956 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 12 |
Hb_000077_280 |
0.1040295432 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_020738_010 |
0.1113123609 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 14 |
Hb_007357_010 |
0.1124061607 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_007123_010 |
0.1154249591 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 16 |
Hb_000996_070 |
0.1170702363 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000032_420 |
0.117415408 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 18 |
Hb_000012_350 |
0.117730663 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 19 |
Hb_000928_280 |
0.1179484886 |
- |
- |
hypothetical protein VITISV_005300 [Vitis vinifera] |
| 20 |
Hb_000185_130 |
0.1182346026 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |