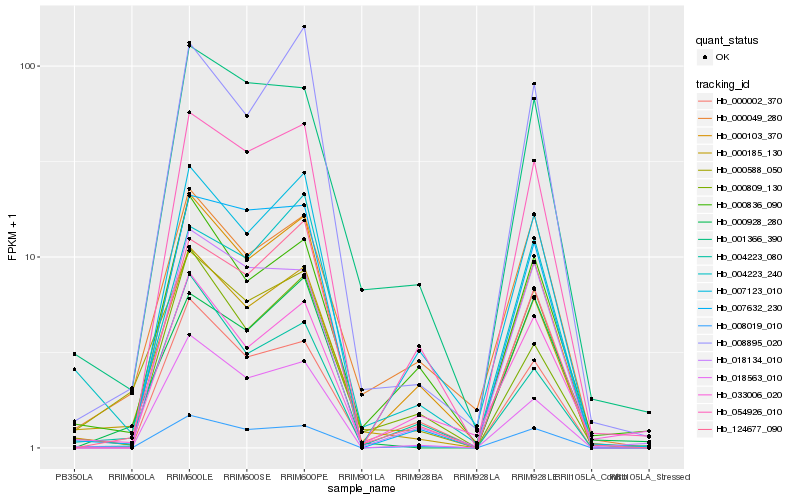
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000185_130 |
0.0 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |
| 2 |
Hb_000588_050 |
0.0838561247 |
transcription factor |
TF Family: Trihelix |
hypothetical protein POPTR_0008s02580g [Populus trichocarpa] |
| 3 |
Hb_000103_370 |
0.1147420222 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 4 |
Hb_008895_020 |
0.1182346026 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 5 |
Hb_001366_390 |
0.1204103951 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 6 |
Hb_000836_090 |
0.1261137187 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 7 |
Hb_124677_090 |
0.1319506063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_054926_010 |
0.1348021884 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 9 |
Hb_000809_130 |
0.1351919932 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000049_280 |
0.1362152571 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 11 |
Hb_008019_010 |
0.1385551473 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 12 |
Hb_000002_370 |
0.1412615111 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 13 |
Hb_000928_280 |
0.1421312375 |
- |
- |
hypothetical protein VITISV_005300 [Vitis vinifera] |
| 14 |
Hb_033006_020 |
0.1439620765 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_007632_230 |
0.1440621502 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 16 |
Hb_018134_010 |
0.1444443075 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007123_010 |
0.1448451153 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 18 |
Hb_004223_080 |
0.1459970298 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_004223_240 |
0.1464377827 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 20 |
Hb_018563_010 |
0.1476666312 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |