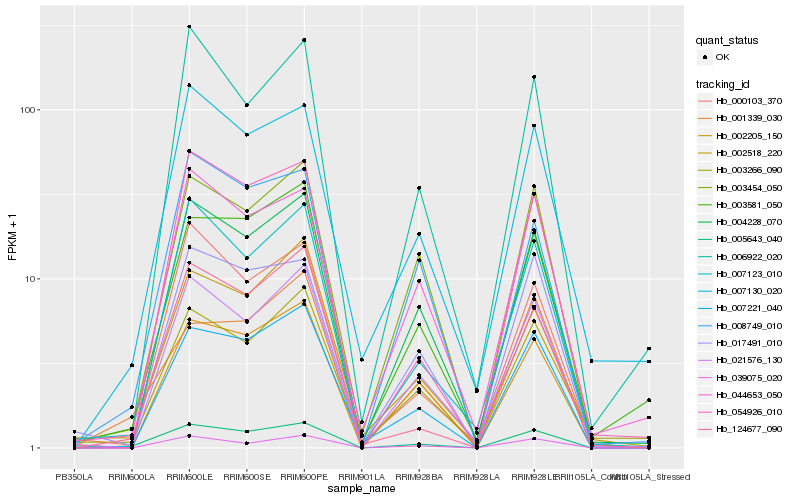
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005643_040 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_004228_070 |
0.110835034 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 3 |
Hb_007221_040 |
0.1153571011 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 4 |
Hb_003454_050 |
0.1188630945 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 5 |
Hb_021576_130 |
0.1200852775 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007123_010 |
0.1235279543 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 7 |
Hb_007130_020 |
0.125377551 |
- |
- |
- |
| 8 |
Hb_002205_150 |
0.1266244344 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 9 |
Hb_054926_010 |
0.1270803273 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 10 |
Hb_017491_010 |
0.1277609956 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003581_050 |
0.1280467872 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 12 |
Hb_008749_010 |
0.1286839043 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 13 |
Hb_044653_050 |
0.1299750016 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001339_030 |
0.1307968847 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 15 |
Hb_039075_020 |
0.1313296565 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 16 |
Hb_003266_090 |
0.134821068 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 17 |
Hb_124677_090 |
0.1349995245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006922_020 |
0.1386652086 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 19 |
Hb_000103_370 |
0.141115617 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 20 |
Hb_002518_220 |
0.1413462228 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |