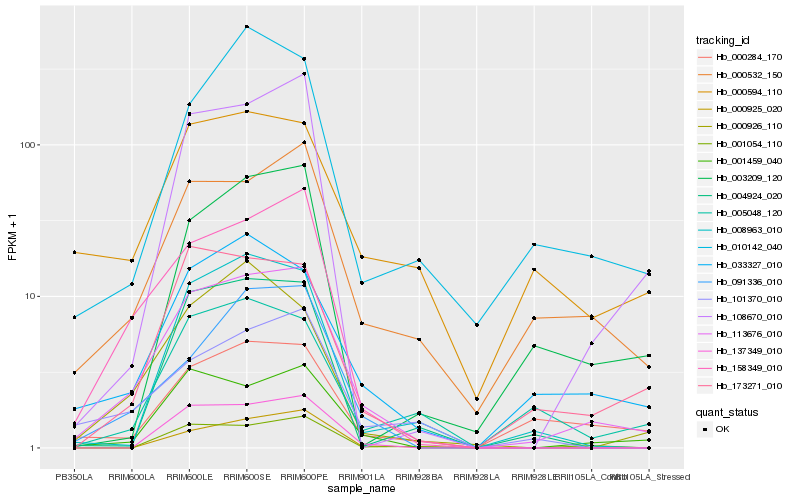
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113676_010 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 2 |
Hb_001459_040 |
0.1425324652 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 3 |
Hb_158349_010 |
0.1504304296 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 4 |
Hb_000532_150 |
0.1531125743 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 5 |
Hb_033327_010 |
0.1631581691 |
- |
- |
SYM8 [Pisum sativum] |
| 6 |
Hb_173271_010 |
0.1639471574 |
- |
- |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 7 |
Hb_108670_010 |
0.1765959386 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 8 |
Hb_137349_010 |
0.1815361523 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 9 |
Hb_001054_110 |
0.1823882549 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 10 |
Hb_091336_010 |
0.1827328879 |
- |
- |
- |
| 11 |
Hb_101370_010 |
0.1831397468 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_008963_010 |
0.1854373243 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000284_170 |
0.1855437652 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000926_110 |
0.1925227989 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 15 |
Hb_000594_110 |
0.1932236084 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 16 |
Hb_005048_120 |
0.1948010548 |
- |
- |
- |
| 17 |
Hb_000925_020 |
0.1979579255 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 18 |
Hb_004924_020 |
0.1981995731 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_010142_040 |
0.1985516236 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 20 |
Hb_003209_120 |
0.2006477337 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |