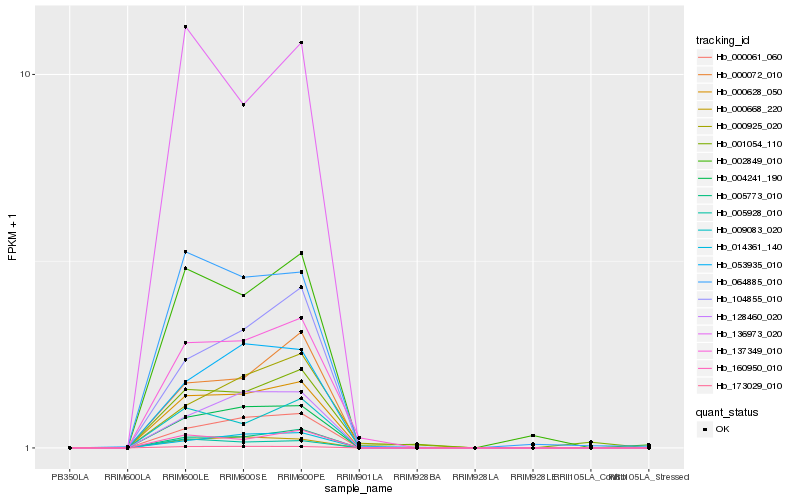
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137349_010 |
0.0 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 2 |
Hb_000628_050 |
0.1017856384 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_004241_190 |
0.1158818313 |
- |
- |
- |
| 4 |
Hb_005773_010 |
0.1172445886 |
- |
- |
- |
| 5 |
Hb_173029_010 |
0.1185114978 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000061_060 |
0.1265169003 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB28 [Arabidopsis thaliana] |
| 7 |
Hb_000668_220 |
0.1350545511 |
- |
- |
PREDICTED: uncharacterized protein LOC105119281 [Populus euphratica] |
| 8 |
Hb_000072_010 |
0.1353631965 |
- |
- |
hypothetical protein B456_006G246700 [Gossypium raimondii] |
| 9 |
Hb_064885_010 |
0.1362826234 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 10 |
Hb_000925_020 |
0.136793784 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 11 |
Hb_160950_010 |
0.137494902 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |
| 12 |
Hb_104855_010 |
0.1380008816 |
- |
- |
- |
| 13 |
Hb_053935_010 |
0.138245032 |
- |
- |
hypothetical protein EUGRSUZ_G02294 [Eucalyptus grandis] |
| 14 |
Hb_136973_020 |
0.1407184447 |
- |
- |
- |
| 15 |
Hb_005928_010 |
0.1422261221 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_002849_010 |
0.1438022505 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 17 |
Hb_009083_020 |
0.1438240241 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 18 |
Hb_128460_020 |
0.144343009 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 19 |
Hb_001054_110 |
0.1452997445 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 20 |
Hb_014361_140 |
0.1457096121 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |