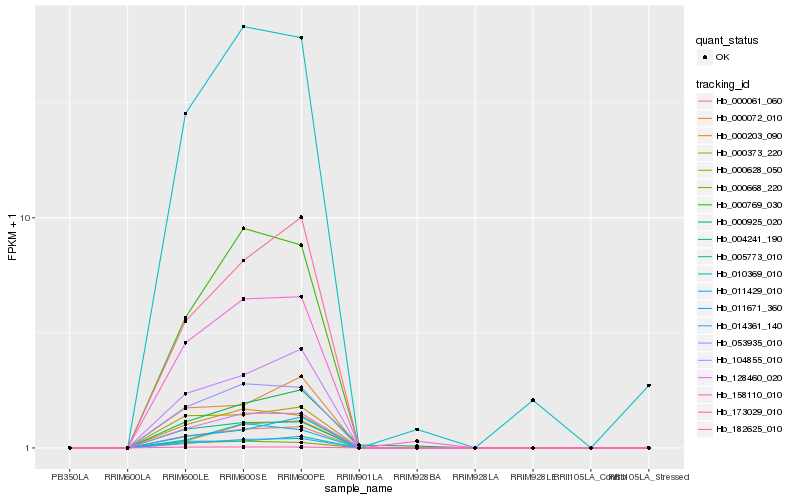
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014361_140 |
0.0 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 2 |
Hb_128460_020 |
0.0239730067 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 3 |
Hb_000061_060 |
0.0295775059 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB28 [Arabidopsis thaliana] |
| 4 |
Hb_053935_010 |
0.0474246318 |
- |
- |
hypothetical protein EUGRSUZ_G02294 [Eucalyptus grandis] |
| 5 |
Hb_104855_010 |
0.0663806776 |
- |
- |
- |
| 6 |
Hb_158110_010 |
0.0665164915 |
- |
- |
hypothetical protein CICLE_v10012017mg [Citrus clementina] |
| 7 |
Hb_004241_190 |
0.0681433604 |
- |
- |
- |
| 8 |
Hb_000769_030 |
0.0725953272 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000373_220 |
0.0730261445 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 10 |
Hb_010369_010 |
0.0869397693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_182625_010 |
0.089858117 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 12 |
Hb_011671_360 |
0.0950528559 |
- |
- |
Hyoscyamine 6-dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_011429_010 |
0.0994162077 |
- |
- |
hypothetical protein POPTR_0188s00230g [Populus trichocarpa] |
| 14 |
Hb_000203_090 |
0.1001334065 |
- |
- |
hypothetical protein POPTR_0013s04080g [Populus trichocarpa] |
| 15 |
Hb_000628_050 |
0.1039052763 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_005773_010 |
0.1046979259 |
- |
- |
- |
| 17 |
Hb_000072_010 |
0.1118415848 |
- |
- |
hypothetical protein B456_006G246700 [Gossypium raimondii] |
| 18 |
Hb_000668_220 |
0.1158261231 |
- |
- |
PREDICTED: uncharacterized protein LOC105119281 [Populus euphratica] |
| 19 |
Hb_173029_010 |
0.122269907 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000925_020 |
0.1279500986 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |