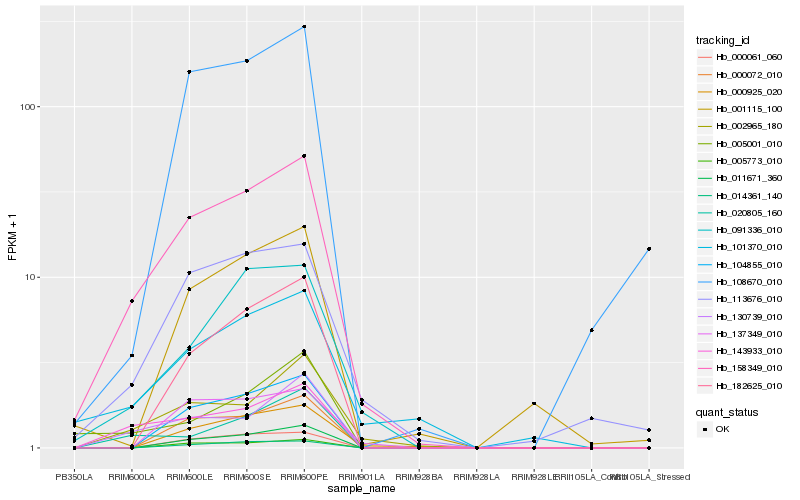
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158349_010 |
0.0 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 2 |
Hb_143933_010 |
0.1271283618 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_002965_180 |
0.1493535148 |
- |
- |
AMSH-like ubiquitin thioesterase 3 [Morus notabilis] |
| 4 |
Hb_113676_010 |
0.1504304296 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 5 |
Hb_091336_010 |
0.1505528524 |
- |
- |
- |
| 6 |
Hb_130739_010 |
0.158230221 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_101370_010 |
0.1646946498 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_020805_160 |
0.1855736827 |
- |
- |
- |
| 9 |
Hb_104855_010 |
0.187414531 |
- |
- |
- |
| 10 |
Hb_011671_360 |
0.191374337 |
- |
- |
Hyoscyamine 6-dioxygenase, putative [Ricinus communis] |
| 11 |
Hb_000072_010 |
0.1926005889 |
- |
- |
hypothetical protein B456_006G246700 [Gossypium raimondii] |
| 12 |
Hb_000925_020 |
0.1942239401 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 13 |
Hb_005773_010 |
0.1949695467 |
- |
- |
- |
| 14 |
Hb_182625_010 |
0.1978916967 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 15 |
Hb_000061_060 |
0.1981616967 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB28 [Arabidopsis thaliana] |
| 16 |
Hb_005001_010 |
0.1996172694 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 17 |
Hb_014361_140 |
0.1999469934 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 18 |
Hb_108670_010 |
0.2001390236 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 19 |
Hb_137349_010 |
0.2047280129 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 20 |
Hb_001115_100 |
0.2074247294 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |