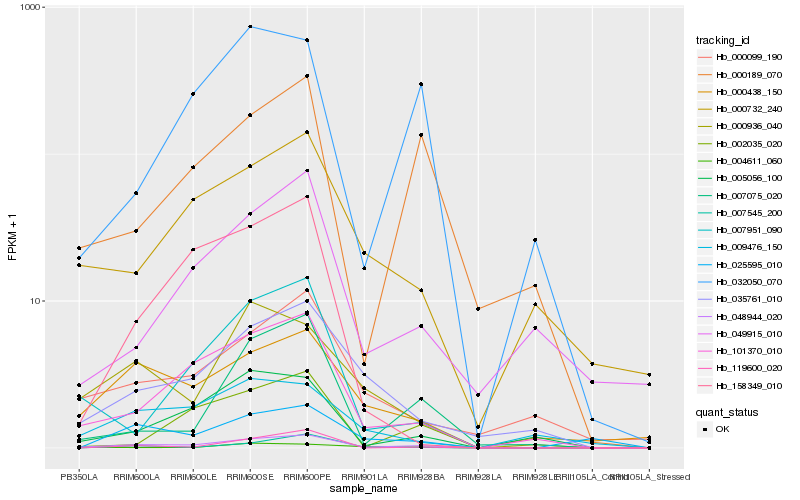
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048944_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s002002g, partial [Populus trichocarpa] |
| 2 |
Hb_101370_010 |
0.160199635 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_035761_010 |
0.2003594967 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 4 |
Hb_000438_150 |
0.203257701 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 5 |
Hb_005056_100 |
0.2110215274 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 6 |
Hb_000099_190 |
0.2174038559 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 7 |
Hb_000189_070 |
0.2176708752 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 8 |
Hb_000732_240 |
0.2178543289 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 9 |
Hb_007951_090 |
0.2224075606 |
- |
- |
hypothetical protein POPTR_0003s06080g [Populus trichocarpa] |
| 10 |
Hb_000936_040 |
0.2241427291 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 11 |
Hb_007075_020 |
0.2265137285 |
transcription factor |
TF Family: TRAF |
ankyrin repeat family protein [Populus trichocarpa] |
| 12 |
Hb_002035_020 |
0.2280267409 |
transcription factor |
TF Family: TRAF |
hypothetical protein B456_011G050200 [Gossypium raimondii] |
| 13 |
Hb_032050_070 |
0.2311305168 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 14 |
Hb_007545_200 |
0.2341686612 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 15 |
Hb_004611_060 |
0.2355838044 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 16 |
Hb_025595_010 |
0.2392269971 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 17 |
Hb_049915_010 |
0.2409586181 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 18 |
Hb_158349_010 |
0.2415029508 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 19 |
Hb_119600_020 |
0.2417334063 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_009476_150 |
0.2440448353 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |