| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
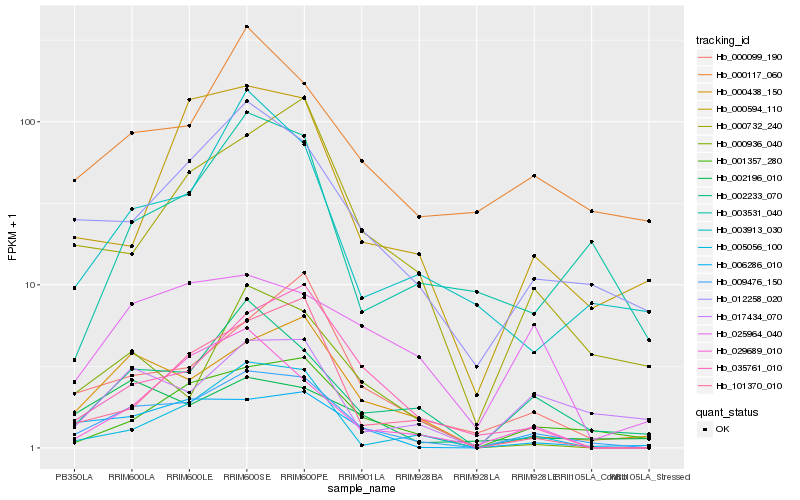
Hb_009476_150 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 2 |
Hb_000438_150 |
0.1693823141 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 3 |
Hb_012258_020 |
0.1753312972 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_002233_070 |
0.1763420199 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001357_280 |
0.1833800248 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 6 |
Hb_006286_010 |
0.1898436114 |
- |
- |
hypothetical protein PRUPE_ppa017148mg [Prunus persica] |
| 7 |
Hb_000732_240 |
0.1937364225 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 8 |
Hb_005056_100 |
0.2092380459 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 9 |
Hb_000099_190 |
0.2094547177 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 10 |
Hb_035761_010 |
0.2099779094 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 11 |
Hb_000936_040 |
0.2115663242 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 12 |
Hb_000117_060 |
0.2179716455 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 13 |
Hb_029689_010 |
0.2180743882 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_101370_010 |
0.2180853374 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_000594_110 |
0.2184496138 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 16 |
Hb_017434_070 |
0.2209297896 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_002196_010 |
0.2248535077 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: proline-rich receptor-like protein kinase PERK1 [Jatropha curcas] |
| 18 |
Hb_003913_030 |
0.2272057627 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 19 |
Hb_025964_040 |
0.2304386136 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 20 |
Hb_003531_040 |
0.2322124069 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |