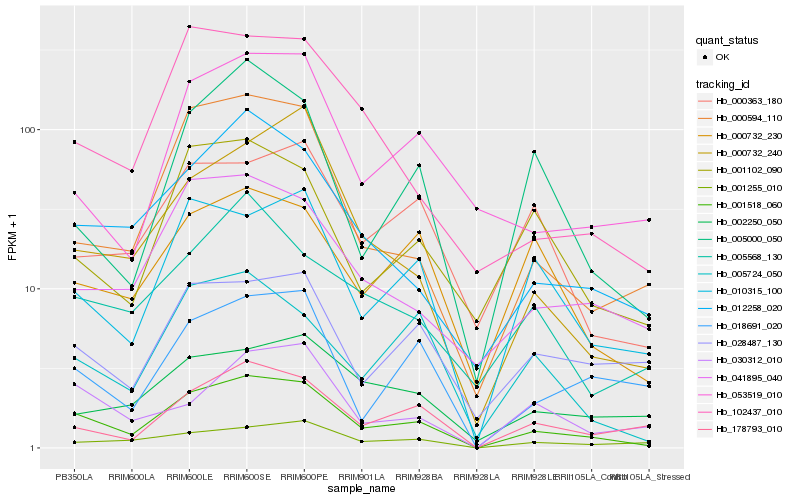
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001518_060 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 2 |
Hb_012258_020 |
0.1631541918 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_010315_100 |
0.1663634278 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 4 |
Hb_178793_010 |
0.1709737521 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Nelumbo nucifera] |
| 5 |
Hb_005724_050 |
0.1739500271 |
- |
- |
- |
| 6 |
Hb_041895_040 |
0.1740158938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000594_110 |
0.1754315579 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 8 |
Hb_001255_010 |
0.1792713962 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Tarenaya hassleriana] |
| 9 |
Hb_000732_230 |
0.1798015869 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_018691_020 |
0.1822695078 |
- |
- |
- |
| 11 |
Hb_028487_130 |
0.1833835563 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 12 |
Hb_001102_090 |
0.1856669908 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 13 |
Hb_102437_010 |
0.1880005996 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_030312_010 |
0.1896264333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005568_130 |
0.1897620569 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_005000_050 |
0.1898517614 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 17 |
Hb_002250_050 |
0.1902180148 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 18 |
Hb_000363_180 |
0.1903015835 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 19 |
Hb_000732_240 |
0.1915619028 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 20 |
Hb_053519_010 |
0.1915695914 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |