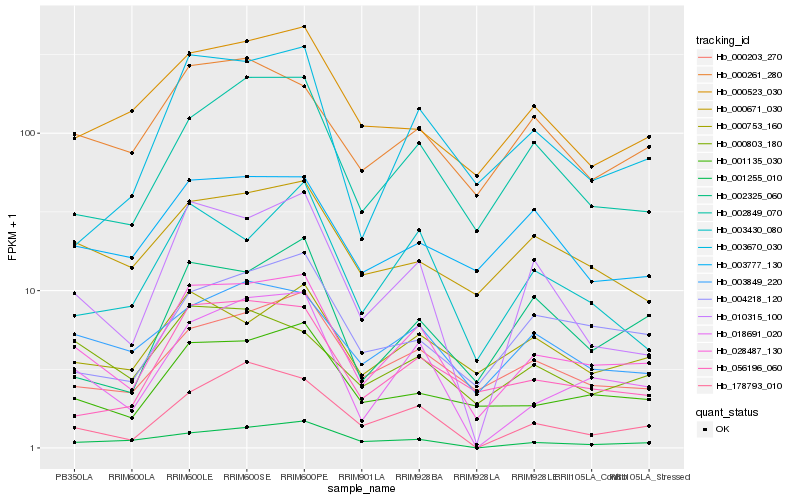
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028487_130 |
0.0 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 2 |
Hb_001135_030 |
0.1221057859 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 3 |
Hb_018691_020 |
0.1235801899 |
- |
- |
- |
| 4 |
Hb_000203_270 |
0.1281096893 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 5 |
Hb_002849_070 |
0.1313123149 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003849_220 |
0.1364421224 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 7 |
Hb_010315_100 |
0.1367524549 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 8 |
Hb_000261_280 |
0.1398591828 |
- |
- |
BnaA10g23810D [Brassica napus] |
| 9 |
Hb_056196_060 |
0.1421654042 |
- |
- |
hexokinase [Manihot esculenta] |
| 10 |
Hb_000671_030 |
0.1476277859 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 11 |
Hb_000803_180 |
0.1489158619 |
- |
- |
hexokinase [Manihot esculenta] |
| 12 |
Hb_004218_120 |
0.1491323868 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000523_030 |
0.1496433114 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 14 |
Hb_178793_010 |
0.1496457259 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Nelumbo nucifera] |
| 15 |
Hb_000753_160 |
0.1503933665 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 16 |
Hb_002325_060 |
0.1505276956 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 17 |
Hb_003670_030 |
0.1510592112 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 18 |
Hb_003430_080 |
0.1515631718 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_001255_010 |
0.1531718781 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Tarenaya hassleriana] |
| 20 |
Hb_003777_130 |
0.1550090433 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |