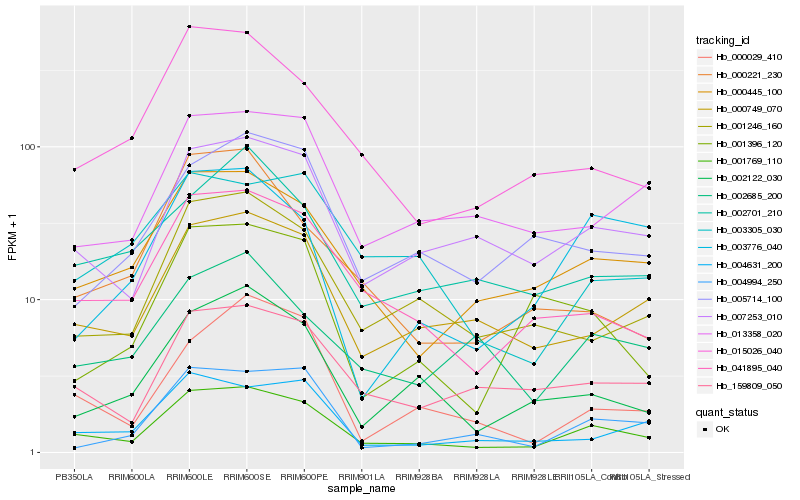
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001769_110 |
0.0 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 2 |
Hb_041895_040 |
0.1330532626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000445_100 |
0.1357221126 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 4 |
Hb_007253_010 |
0.1400021648 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_000749_070 |
0.1463039069 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_159809_050 |
0.150139751 |
- |
- |
AT14A, putative [Ricinus communis] |
| 7 |
Hb_015026_040 |
0.1546200316 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 8 |
Hb_001246_160 |
0.1570025863 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 9 |
Hb_004631_200 |
0.1639140755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002122_030 |
0.1639380783 |
- |
- |
- |
| 11 |
Hb_013358_020 |
0.1688408491 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 12 |
Hb_004994_250 |
0.1689275251 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 13 |
Hb_000029_410 |
0.1757203368 |
- |
- |
AT14A, putative [Ricinus communis] |
| 14 |
Hb_005714_100 |
0.1801928596 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 15 |
Hb_002685_200 |
0.1822714288 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 16 |
Hb_002701_210 |
0.18399647 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 17 |
Hb_001396_120 |
0.1844954588 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 18 |
Hb_003305_030 |
0.1846277697 |
- |
- |
- |
| 19 |
Hb_003776_040 |
0.1883075648 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000221_230 |
0.1883238206 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |