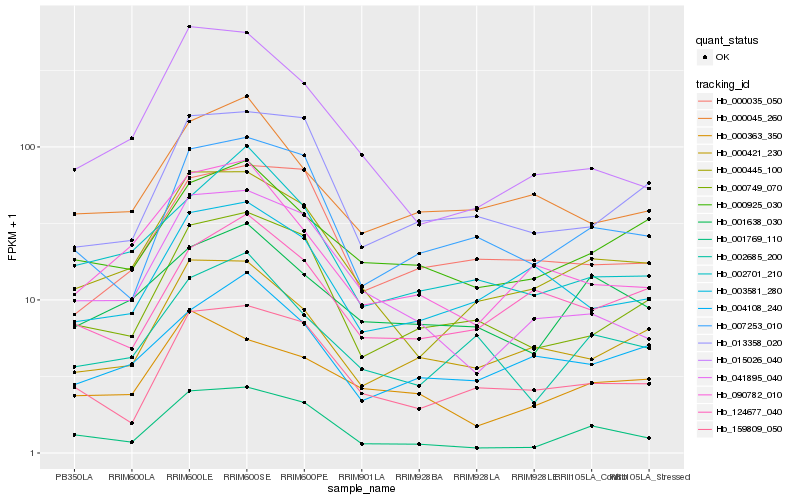
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_100 |
0.0 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 2 |
Hb_015026_040 |
0.1144133971 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 3 |
Hb_159809_050 |
0.1222303968 |
- |
- |
AT14A, putative [Ricinus communis] |
| 4 |
Hb_000421_230 |
0.127640941 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 5 |
Hb_000749_070 |
0.1305274898 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_090782_010 |
0.1312540293 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 7 |
Hb_003581_280 |
0.1345608514 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 8 |
Hb_001769_110 |
0.1357221126 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 9 |
Hb_041895_040 |
0.1367934125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002685_200 |
0.1370089597 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 11 |
Hb_013358_020 |
0.1400896922 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 12 |
Hb_007253_010 |
0.1413234562 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 13 |
Hb_000925_030 |
0.1463393923 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_000035_050 |
0.1491168846 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 15 |
Hb_000363_350 |
0.1496418899 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 16 |
Hb_002701_210 |
0.1517220981 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 17 |
Hb_004108_240 |
0.1521880462 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 18 |
Hb_001638_030 |
0.1522108562 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 19 |
Hb_124677_040 |
0.152322215 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 20 |
Hb_000045_260 |
0.1531816628 |
- |
- |
protein with unknown function [Ricinus communis] |