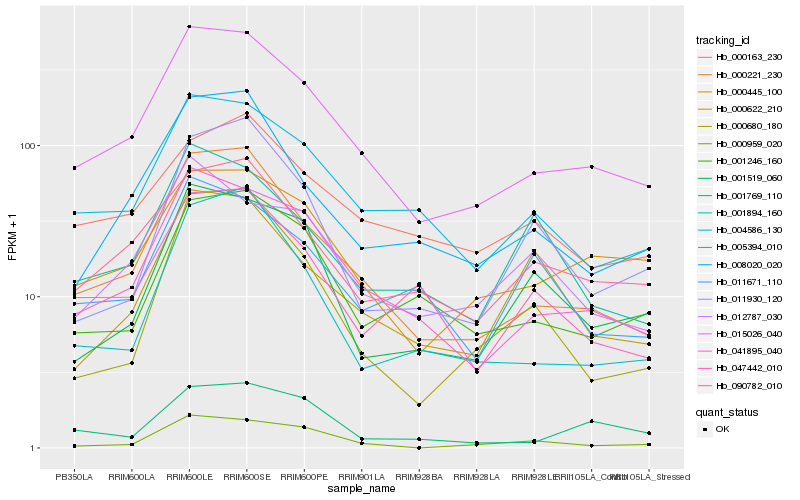
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015026_040 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 2 |
Hb_000221_230 |
0.0731868298 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 3 |
Hb_041895_040 |
0.101333454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005394_010 |
0.1067029539 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 5 |
Hb_000445_100 |
0.1144133971 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 6 |
Hb_090782_010 |
0.1189544188 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 7 |
Hb_008020_020 |
0.1231869388 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001246_160 |
0.1363702718 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 9 |
Hb_047442_010 |
0.1432581978 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 10 |
Hb_004586_130 |
0.1462559673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000163_230 |
0.1464640363 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 12 |
Hb_001519_060 |
0.1474255641 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_012787_030 |
0.1513067787 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 14 |
Hb_001769_110 |
0.1546200316 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 15 |
Hb_000959_020 |
0.1567142944 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_001894_160 |
0.1597650058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011671_110 |
0.1597901756 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 18 |
Hb_000622_210 |
0.1623419891 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_000680_180 |
0.1624730089 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 20 |
Hb_011930_120 |
0.1670322249 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |