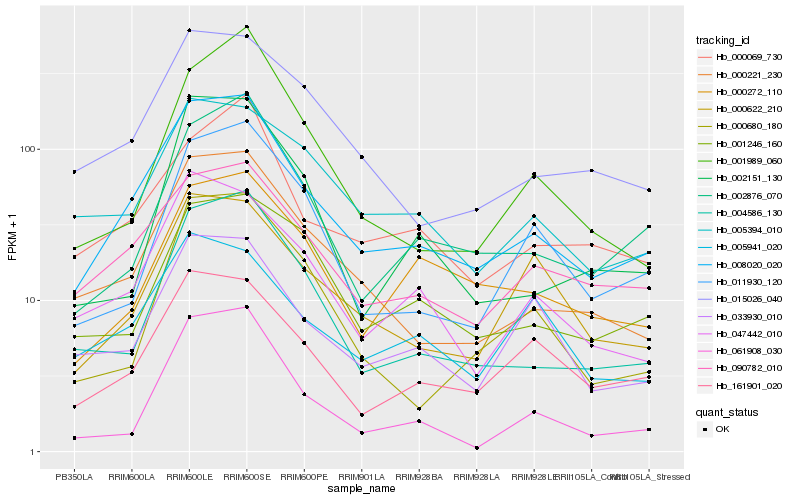
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008020_020 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000221_230 |
0.1075071034 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 3 |
Hb_090782_010 |
0.1159496635 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 4 |
Hb_015026_040 |
0.1231869388 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 5 |
Hb_047442_010 |
0.1279688266 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 6 |
Hb_004586_130 |
0.1282338917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002876_070 |
0.1329838828 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 8 |
Hb_161901_020 |
0.1355162582 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 9 |
Hb_000069_730 |
0.1377480207 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005394_010 |
0.1431973305 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 11 |
Hb_033930_010 |
0.1475666025 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 12 |
Hb_011930_120 |
0.149476512 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000622_210 |
0.1500735647 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_000272_110 |
0.1511629423 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001246_160 |
0.154752803 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 16 |
Hb_000680_180 |
0.1568938287 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 17 |
Hb_002151_130 |
0.1575845318 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 18 |
Hb_061908_030 |
0.1579673517 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 19 |
Hb_005941_020 |
0.1621190341 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 20 |
Hb_001989_060 |
0.1639085871 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |