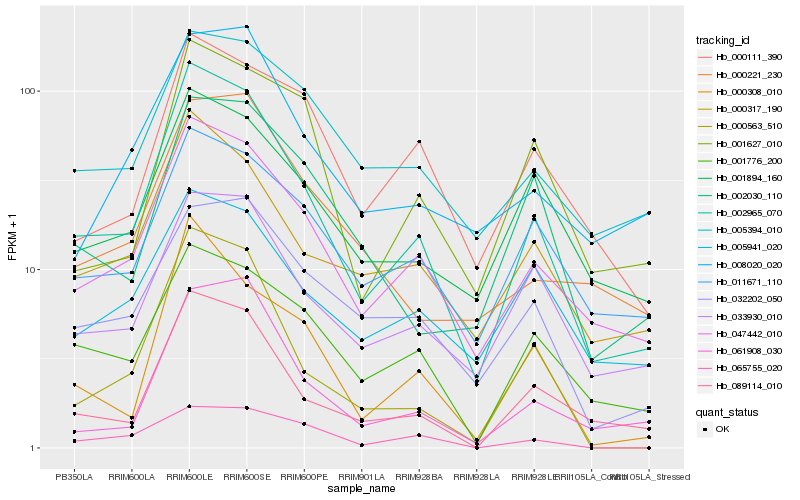
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002965_070 |
0.0 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 2 |
Hb_047442_010 |
0.1007187316 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 3 |
Hb_089114_010 |
0.122780503 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 4 |
Hb_000317_190 |
0.1379628928 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 5 |
Hb_000563_510 |
0.1406761732 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000308_010 |
0.1474878721 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 7 |
Hb_033930_010 |
0.1527691868 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 8 |
Hb_001776_200 |
0.1528600278 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 9 |
Hb_001894_160 |
0.161494403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011671_110 |
0.162035078 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 11 |
Hb_032202_050 |
0.1627217337 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_061908_030 |
0.1630264249 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 13 |
Hb_008020_020 |
0.1721279913 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001627_010 |
0.1727956397 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_000221_230 |
0.173159135 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 16 |
Hb_065755_020 |
0.1755414173 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 17 |
Hb_000111_390 |
0.175650632 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005394_010 |
0.1765885094 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 19 |
Hb_002030_110 |
0.1775573053 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 20 |
Hb_005941_020 |
0.1779841298 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |