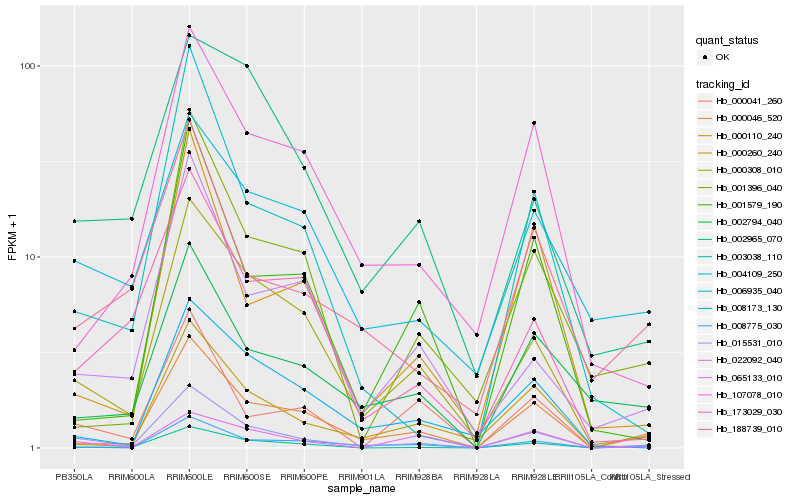
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015531_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636764 [Jatropha curcas] |
| 2 |
Hb_000308_010 |
0.1457757688 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 3 |
Hb_000046_520 |
0.1652167467 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003038_110 |
0.1775469037 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 5 |
Hb_008775_030 |
0.1853077045 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_004109_250 |
0.1932803184 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 7 |
Hb_000041_260 |
0.2012753187 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 8 |
Hb_002794_040 |
0.2021683654 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 9 |
Hb_022092_040 |
0.204353307 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 10 |
Hb_000260_240 |
0.20528186 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 11 |
Hb_008173_130 |
0.208077673 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 12 |
Hb_173029_030 |
0.2096403476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_107078_010 |
0.210580486 |
- |
- |
- |
| 14 |
Hb_065133_010 |
0.212396291 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 15 |
Hb_188739_010 |
0.2141691938 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_006935_040 |
0.2149612873 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 17 |
Hb_000110_240 |
0.215420438 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002965_070 |
0.2161338603 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 19 |
Hb_001579_190 |
0.2168870907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001396_040 |
0.2171764671 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |