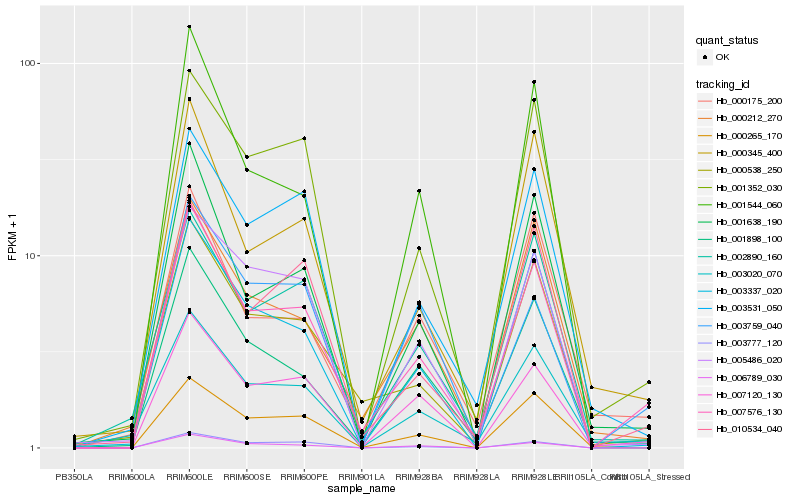
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_130 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 2 |
Hb_007120_130 |
0.1130989236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000538_250 |
0.1223248882 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 4 |
Hb_001638_190 |
0.1274994281 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001898_100 |
0.129665729 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 6 |
Hb_001352_030 |
0.1300547683 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 7 |
Hb_003337_020 |
0.1317904482 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 8 |
Hb_003020_070 |
0.1326335755 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 9 |
Hb_010534_040 |
0.1327049497 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006789_030 |
0.1376634101 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 11 |
Hb_000345_400 |
0.1405958141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000265_170 |
0.1420602282 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_002890_160 |
0.1442292123 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003531_050 |
0.1454391105 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000212_270 |
0.1478378257 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 16 |
Hb_005486_020 |
0.1484250392 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 17 |
Hb_003777_120 |
0.1484705135 |
- |
- |
- |
| 18 |
Hb_000175_200 |
0.1491819099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003759_040 |
0.150379767 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 20 |
Hb_001544_060 |
0.1513139864 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |